Genome assembly: mm10
Demo dataset (BHLHE40 ChIP-seq experiment in MEL [ENCSR000ESH])
| ID | Cell type | TF | Family | Number of input regions with TFBS | CG beta scores in the TFBS | MethMotif logo | Details |
|---|---|---|---|---|---|---|---|
| 1 | MEL | BHLHE40 (Details) | Hairy-related factors | 3614 | 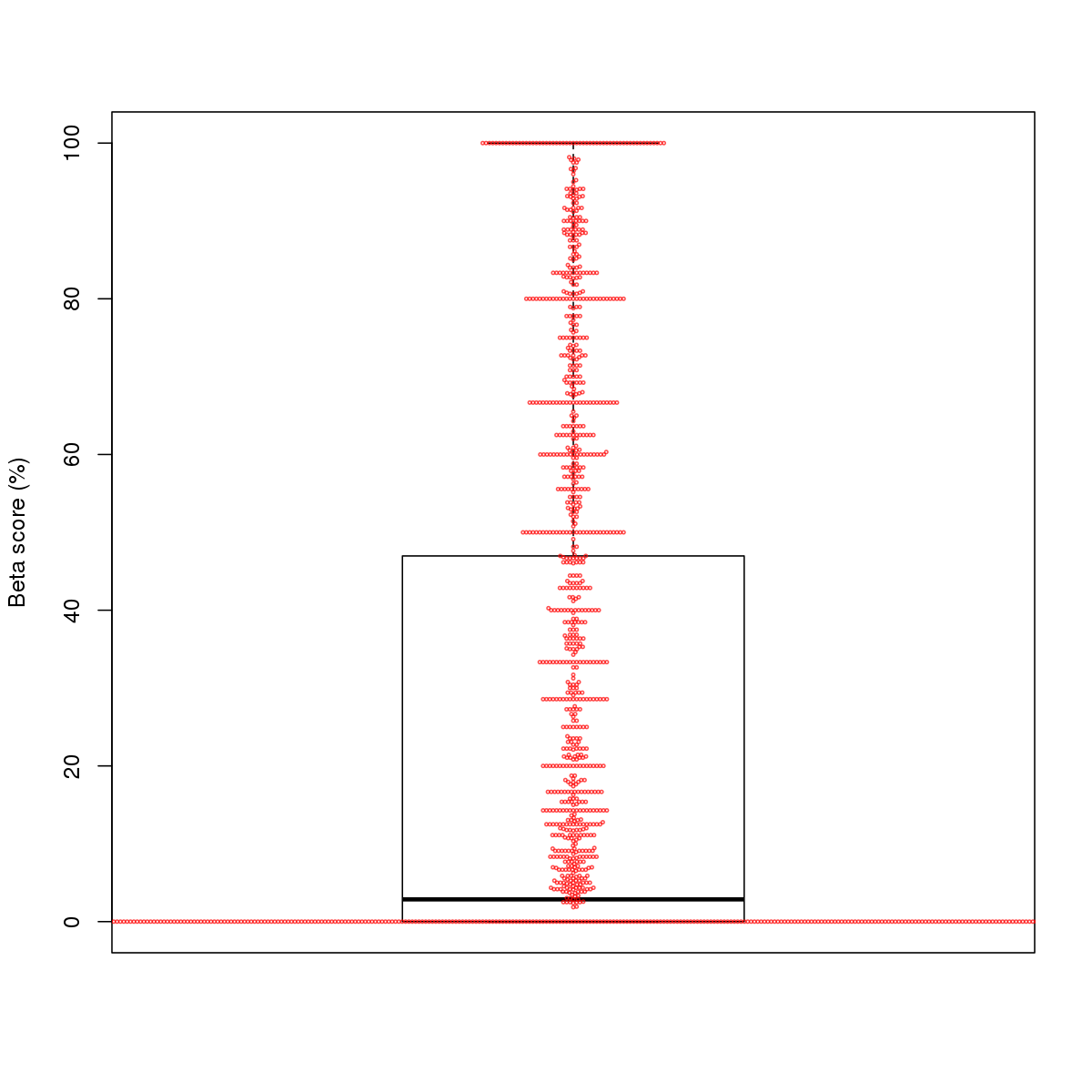 | 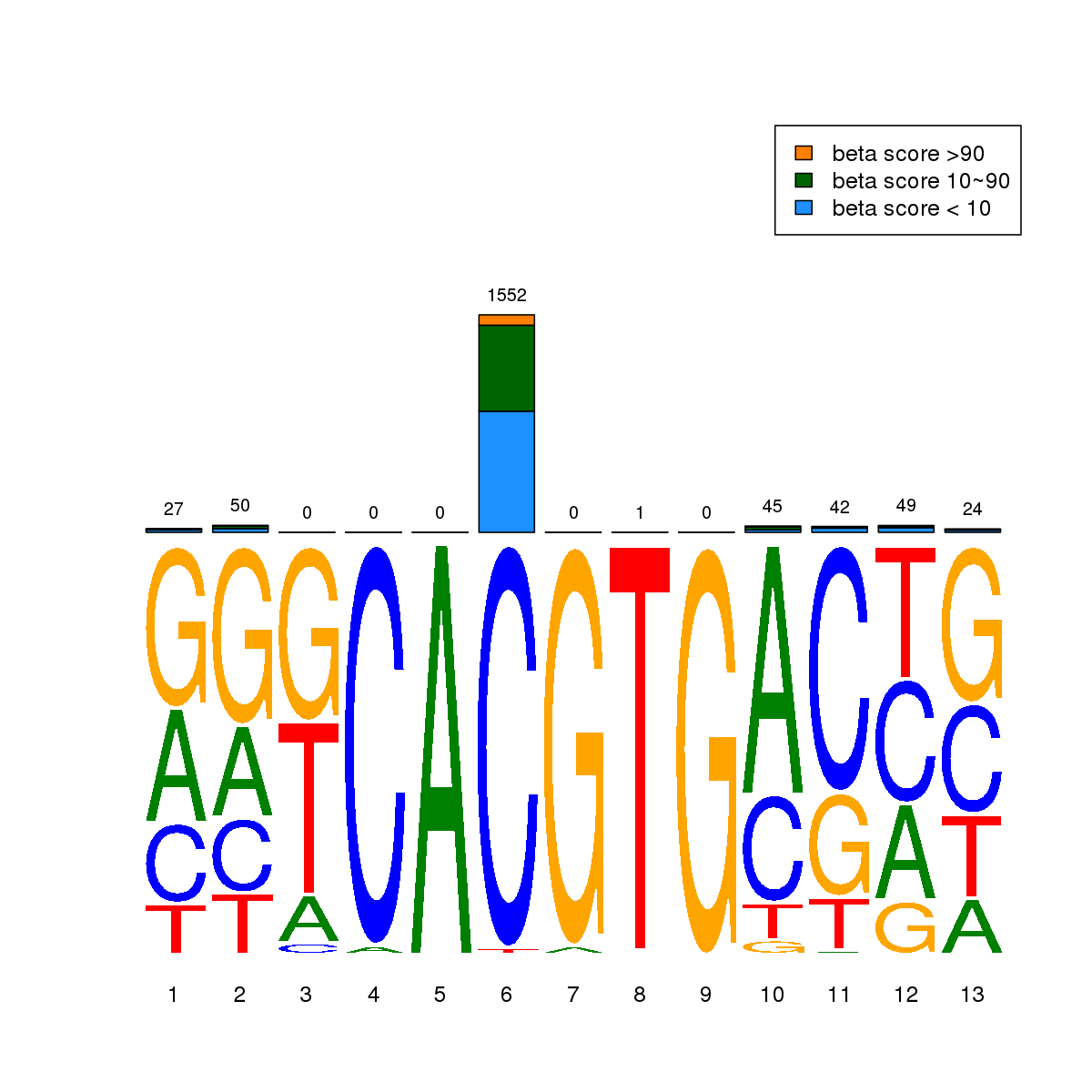 | Regions with BHLHE40 motif: CG methScore in motif: Beta score matrix: |
| 2 | MEL | USF1 (Details) | bHLH-ZIP factors | 1852 | 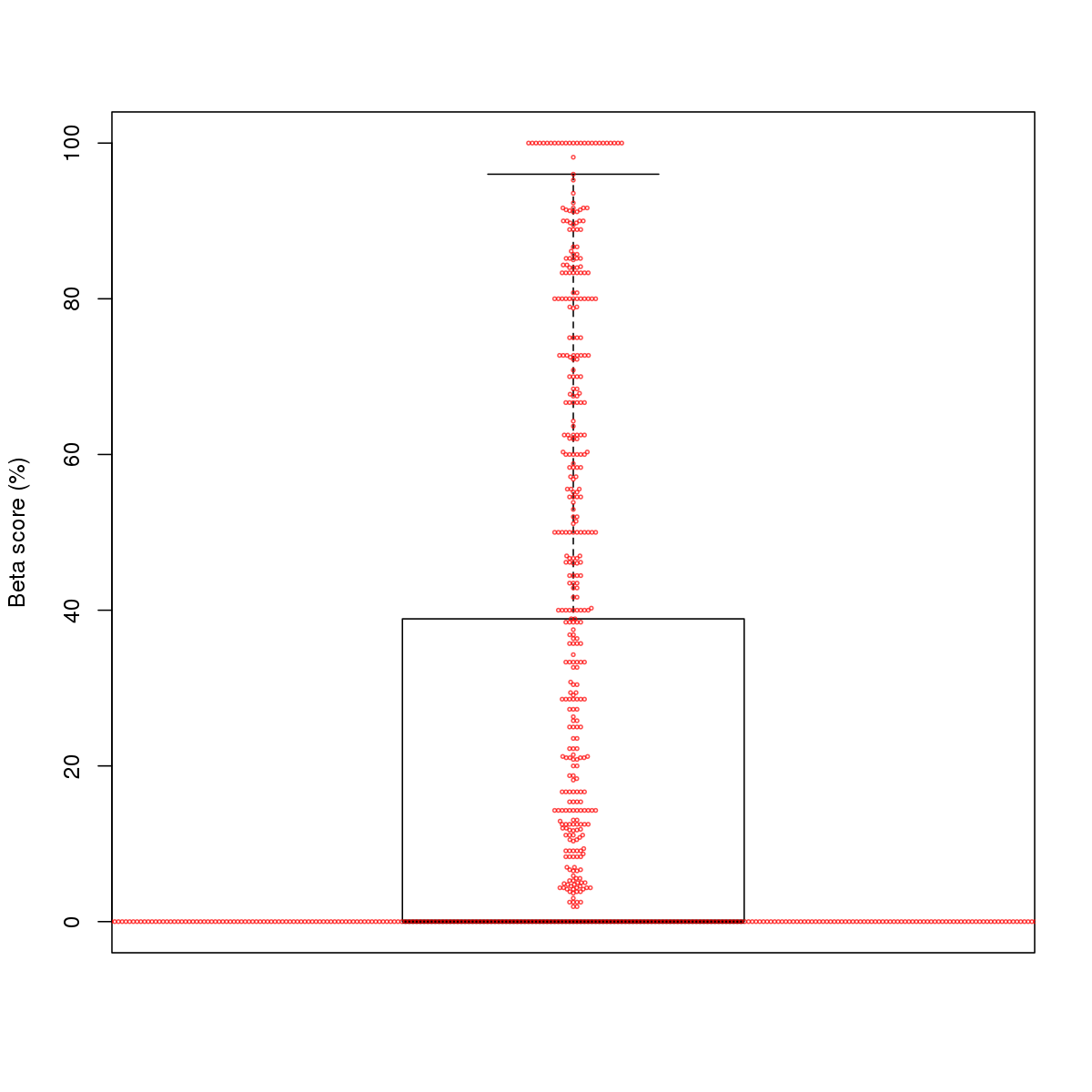 | 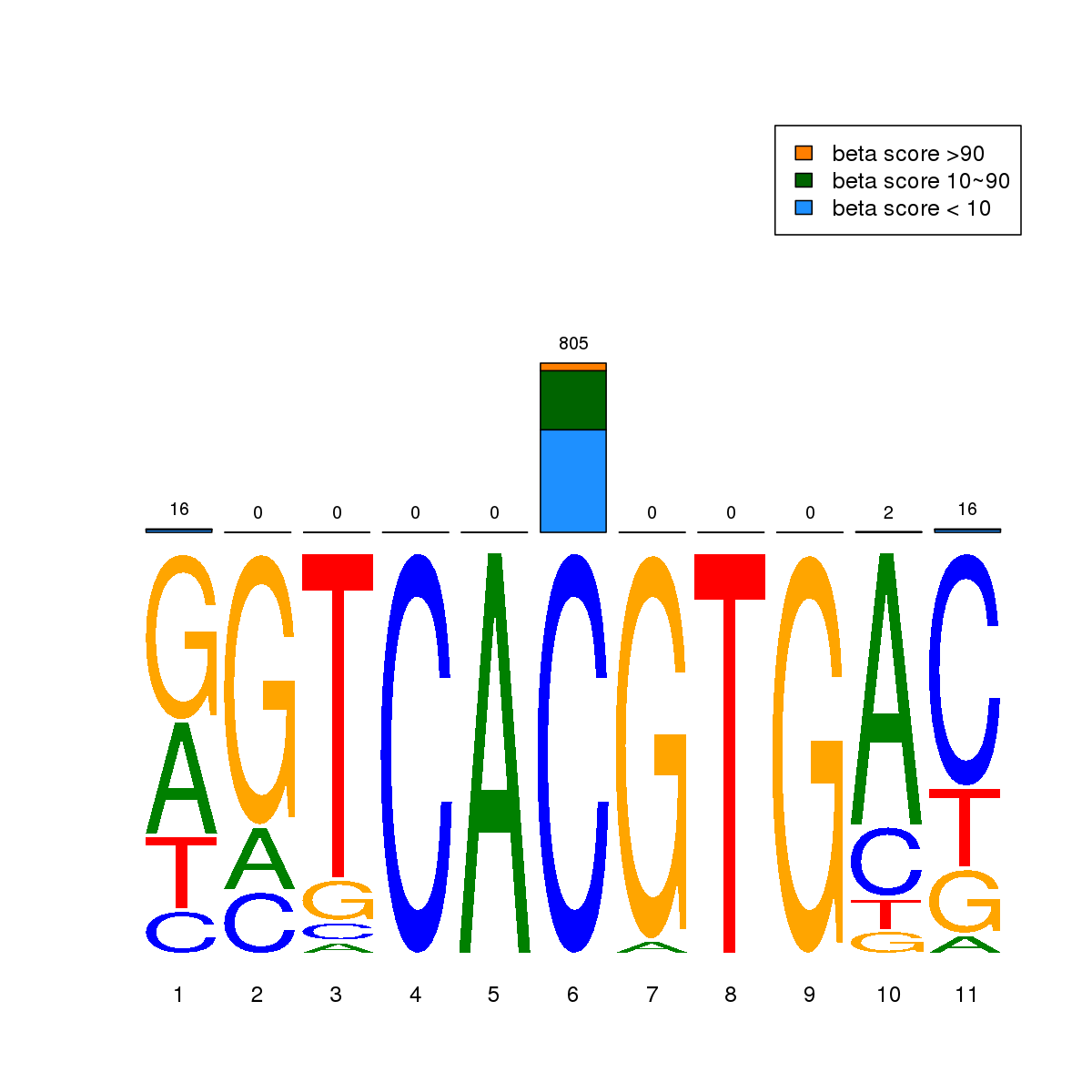 | Regions with USF1 motif: CG methScore in motif: Beta score matrix: |
| 3 | MEL | MAX (Details) | bHLH-ZIP factors | 1020 | 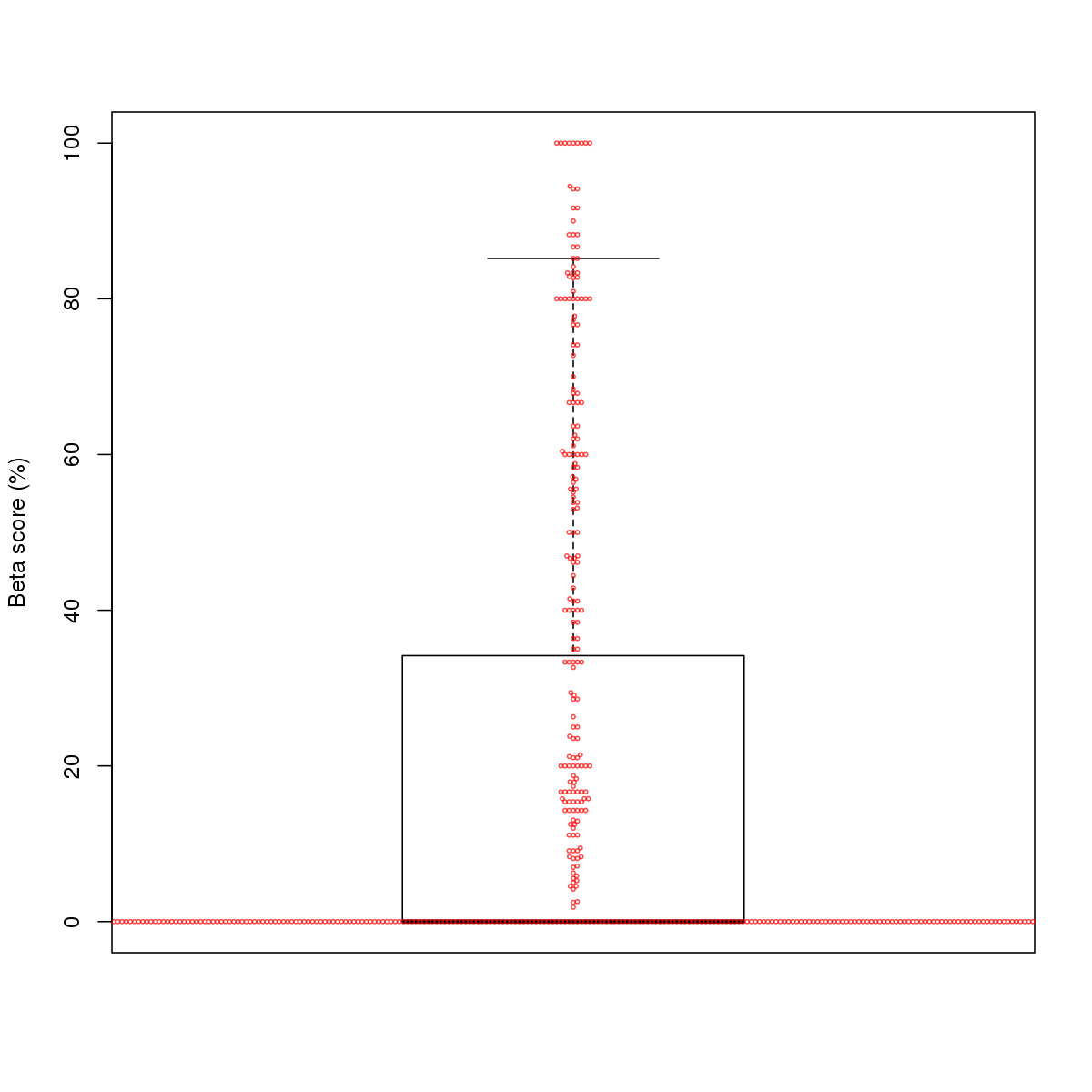 | 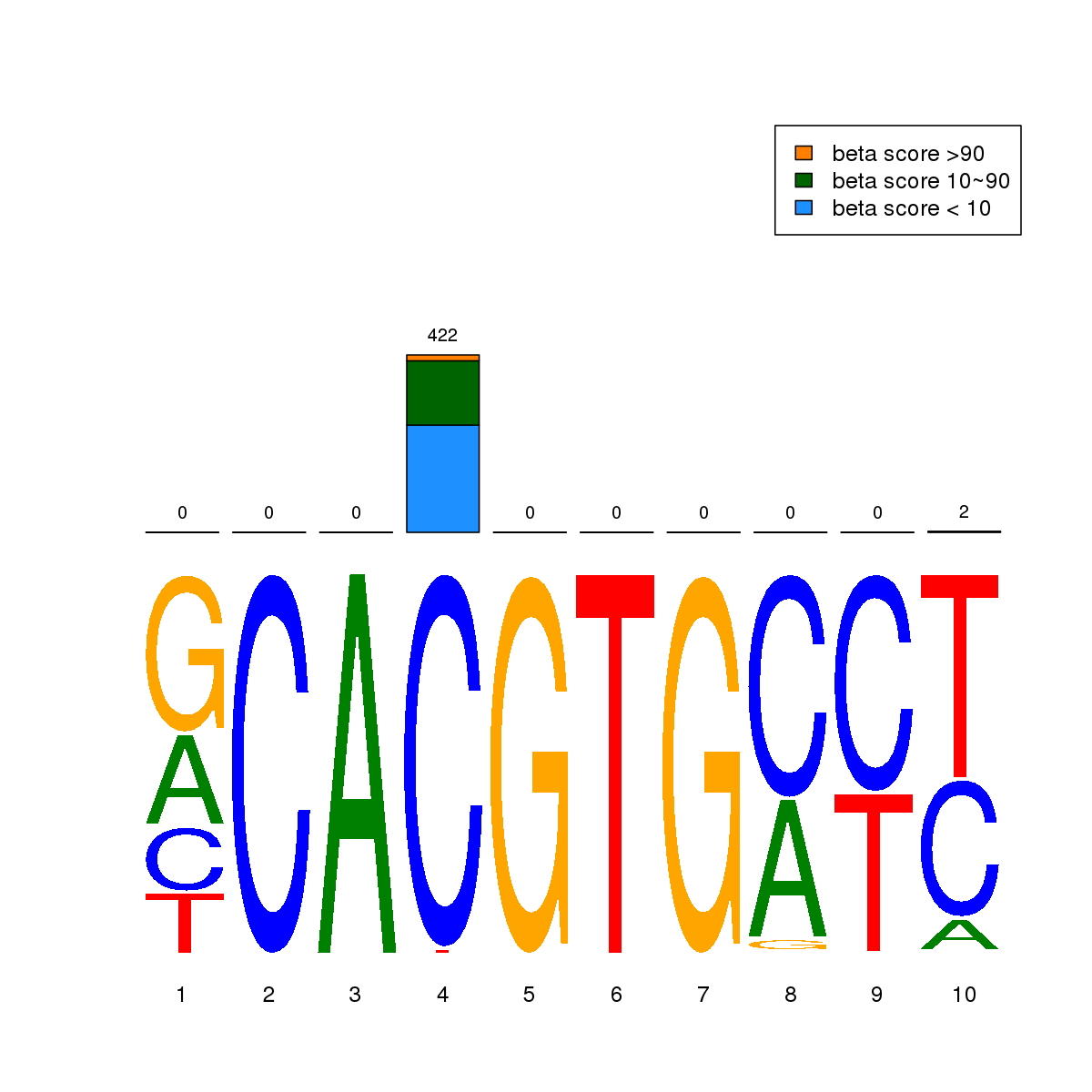 | Regions with MAX motif: CG methScore in motif: Beta score matrix: |
| 4 | MEL | USF2 (Details) | bHLH-ZIP factors | 823 |  | 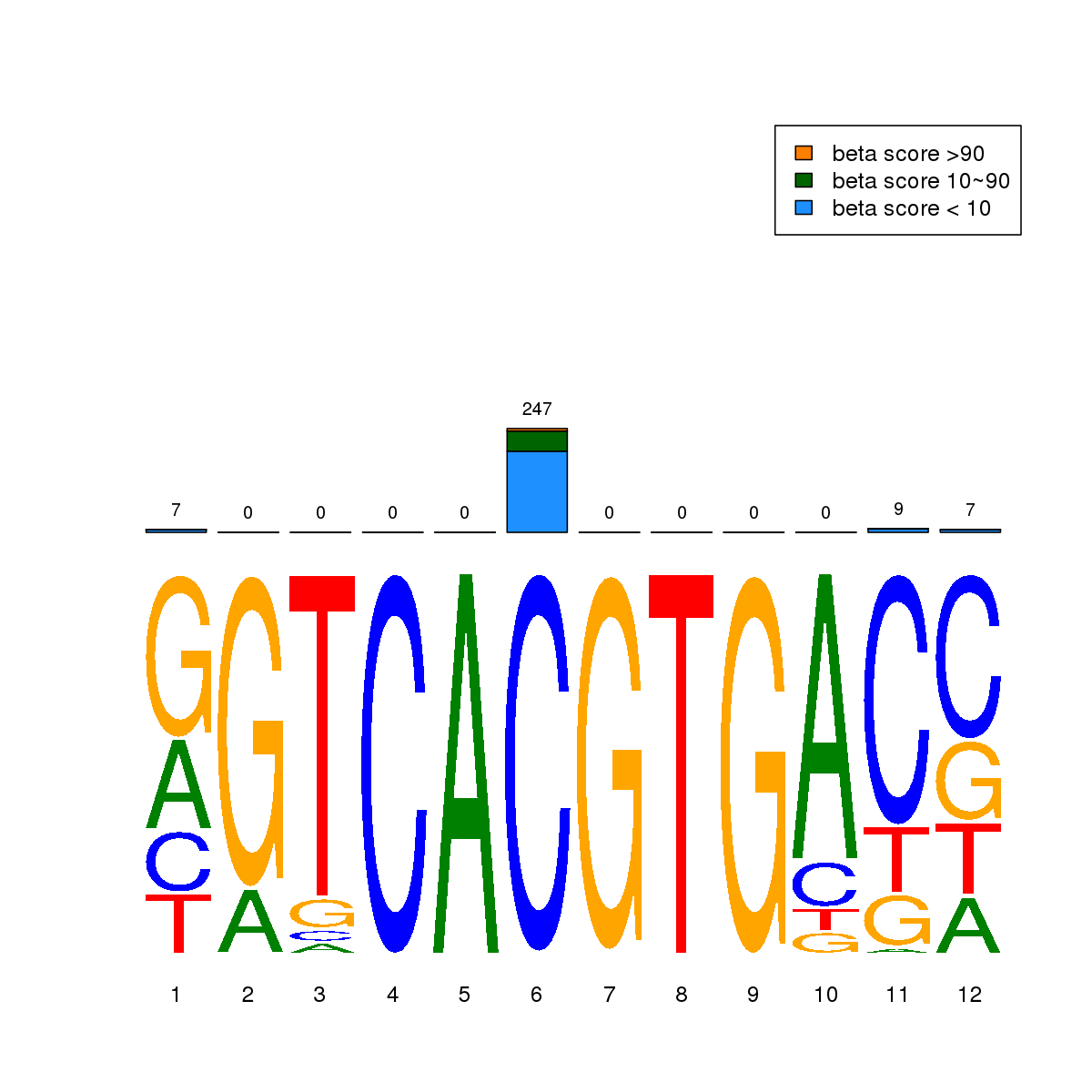 | Regions with USF2 motif: CG methScore in motif: Beta score matrix: |
| 5 | MEL | MXI1 (Details) | bHLH-ZIP factors | 248 | 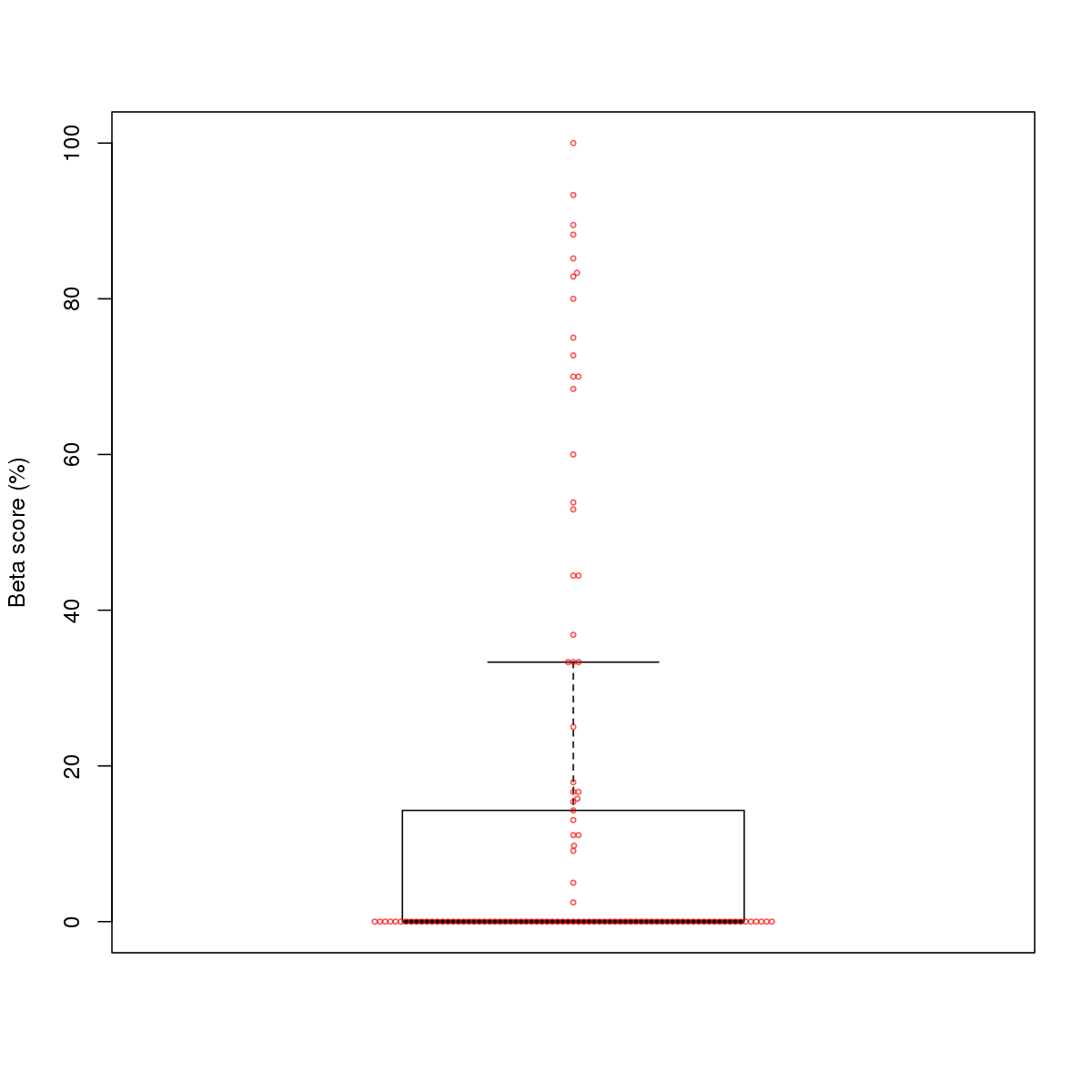 | 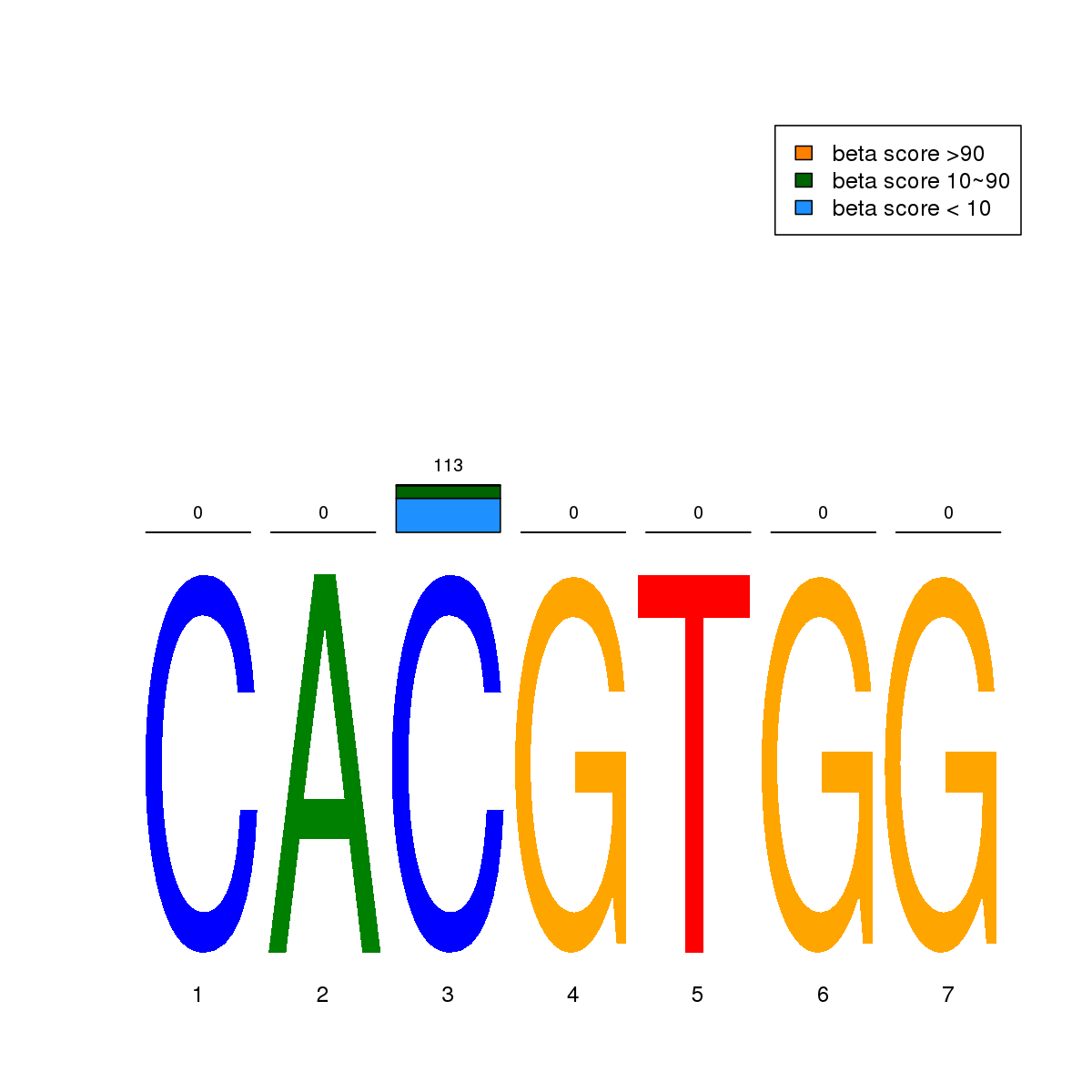 | Regions with MXI1 motif: CG methScore in motif: Beta score matrix: |
| 6 | MEL | MYC (Details) | bHLH-ZIP factors | 105 |  |  | Regions with MYC motif: CG methScore in motif: Beta score matrix: |
| 7 | MEL | NRF1 (Details) | NRF | 32 | 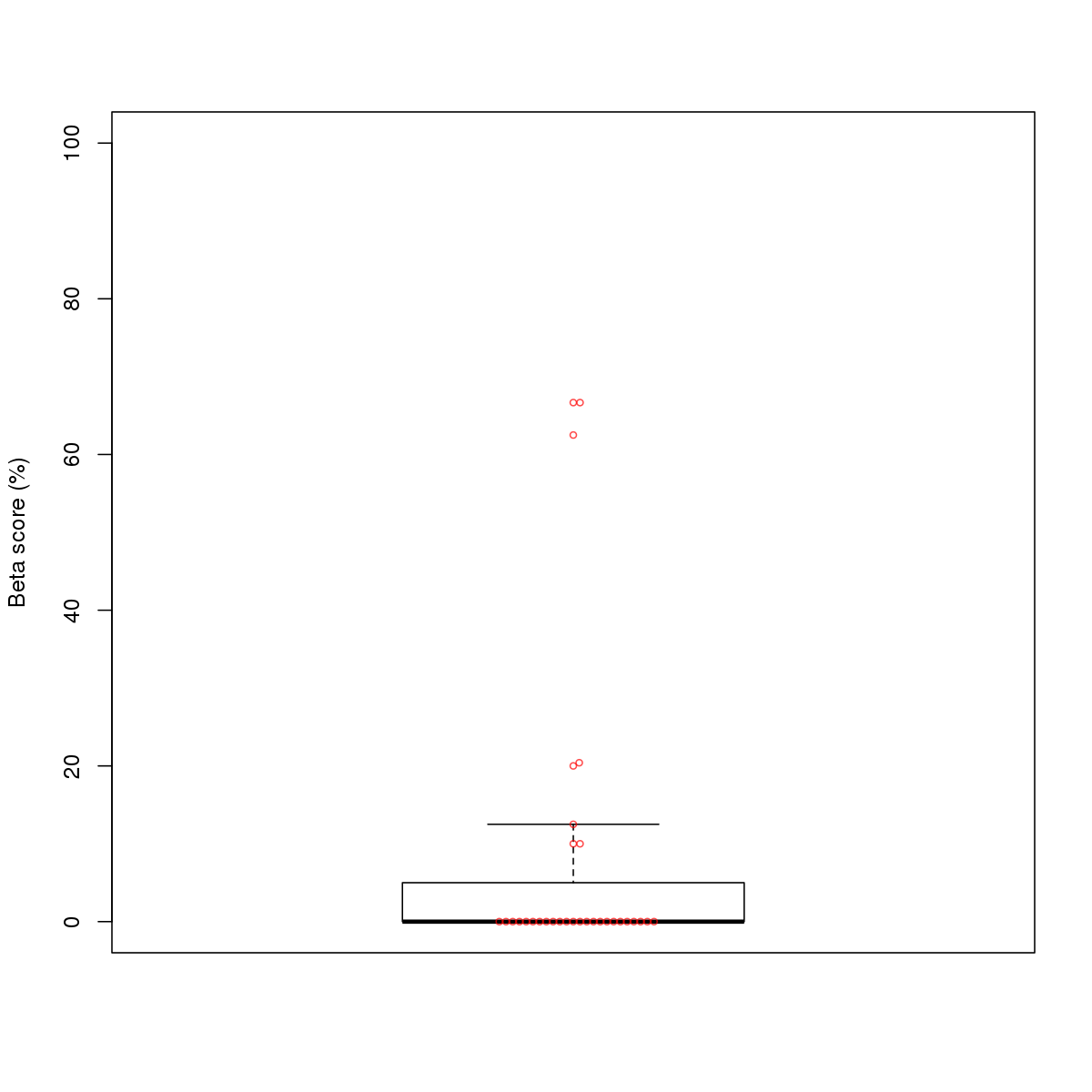 | 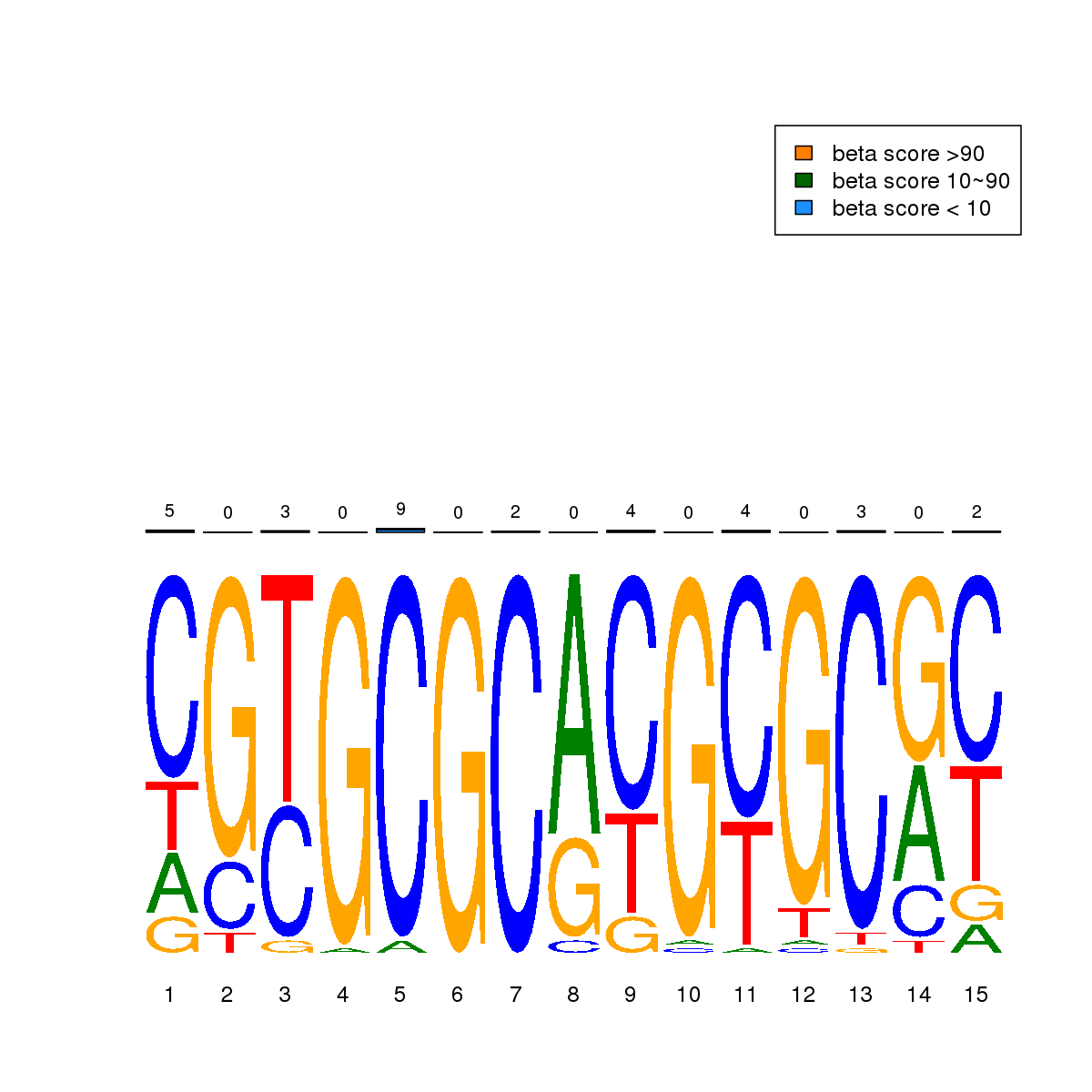 | Regions with NRF1 motif: CG methScore in motif: Beta score matrix: |
| 8 | MEL | GATA1 (Details) | GATA-type zinc fingers | 23 | 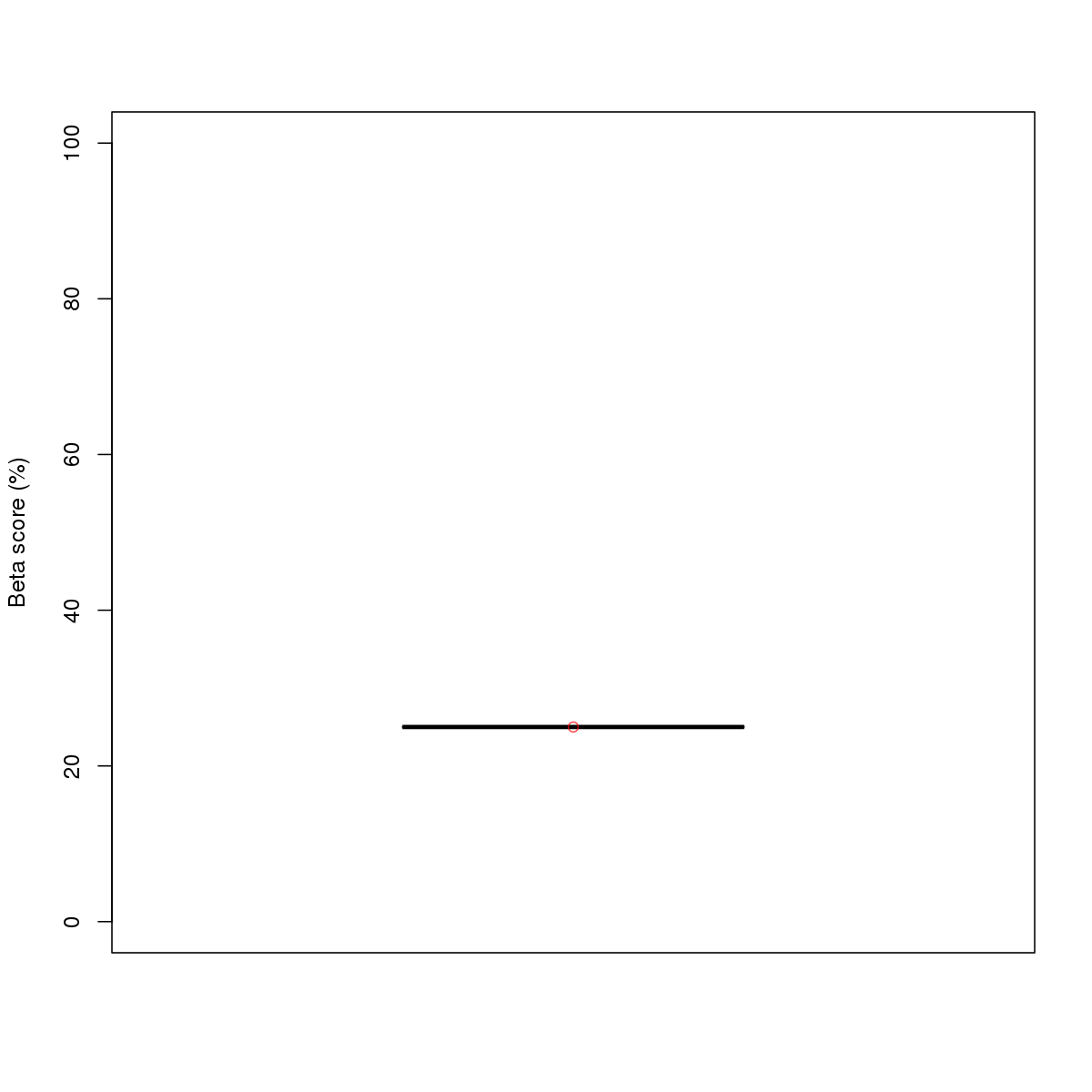 | 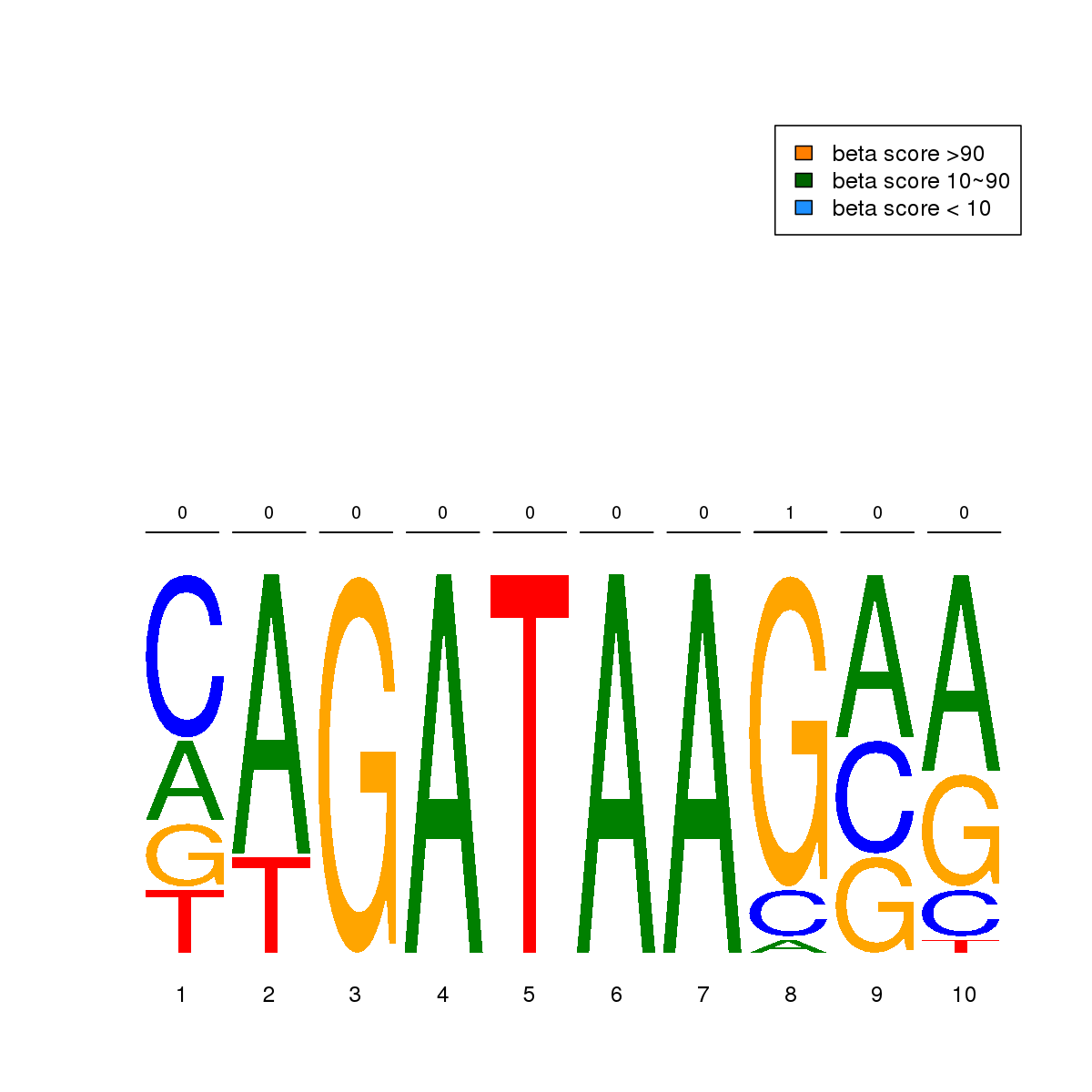 | Regions with GATA1 motif: CG methScore in motif: Beta score matrix: |
| 9 | MEL | TAL1 (Details) | Tal-related factors | 22 | 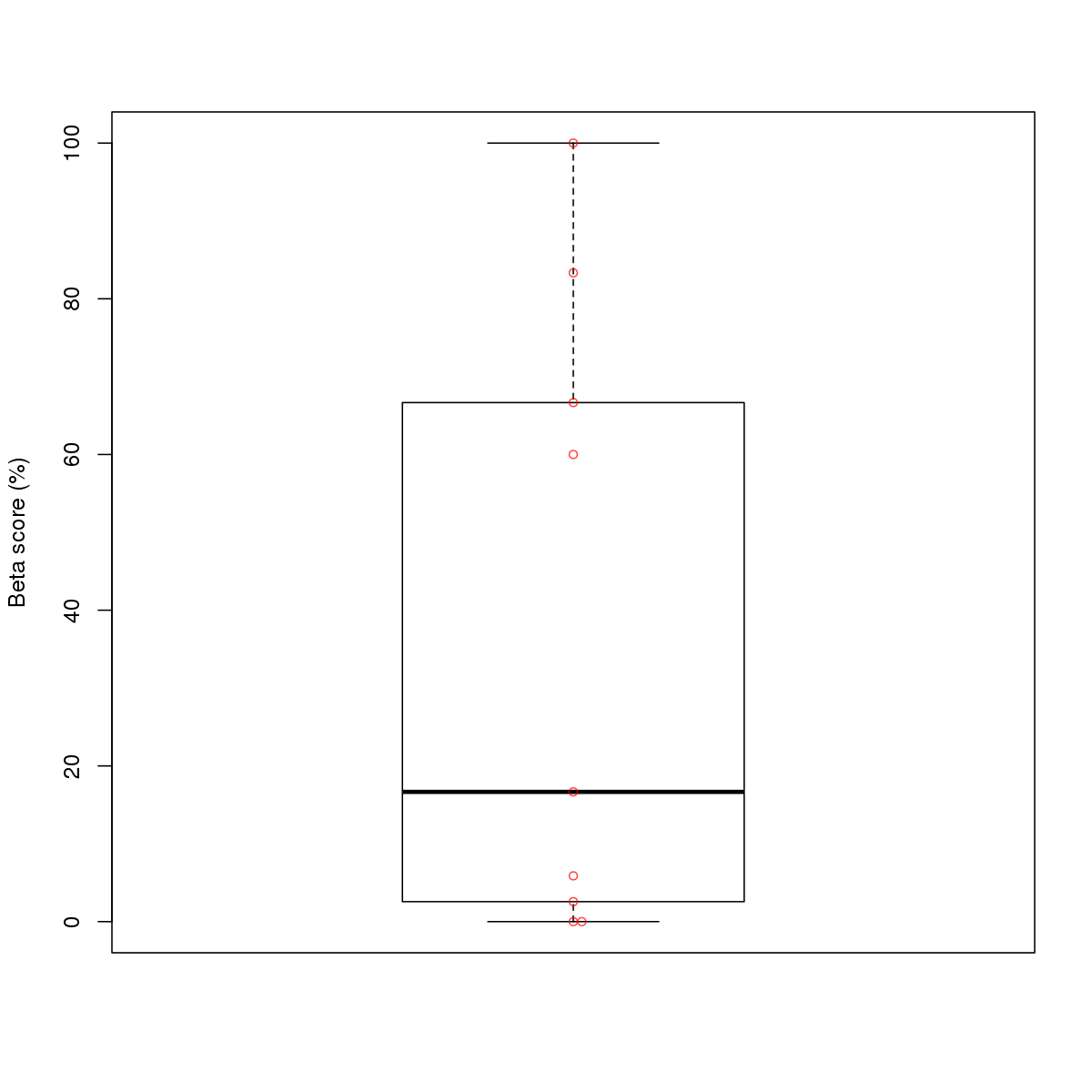 |  | Regions with TAL1 motif: CG methScore in motif: Beta score matrix: |
| 10 | MEL | ELF1 (Details) | Ets-related factors | 20 | 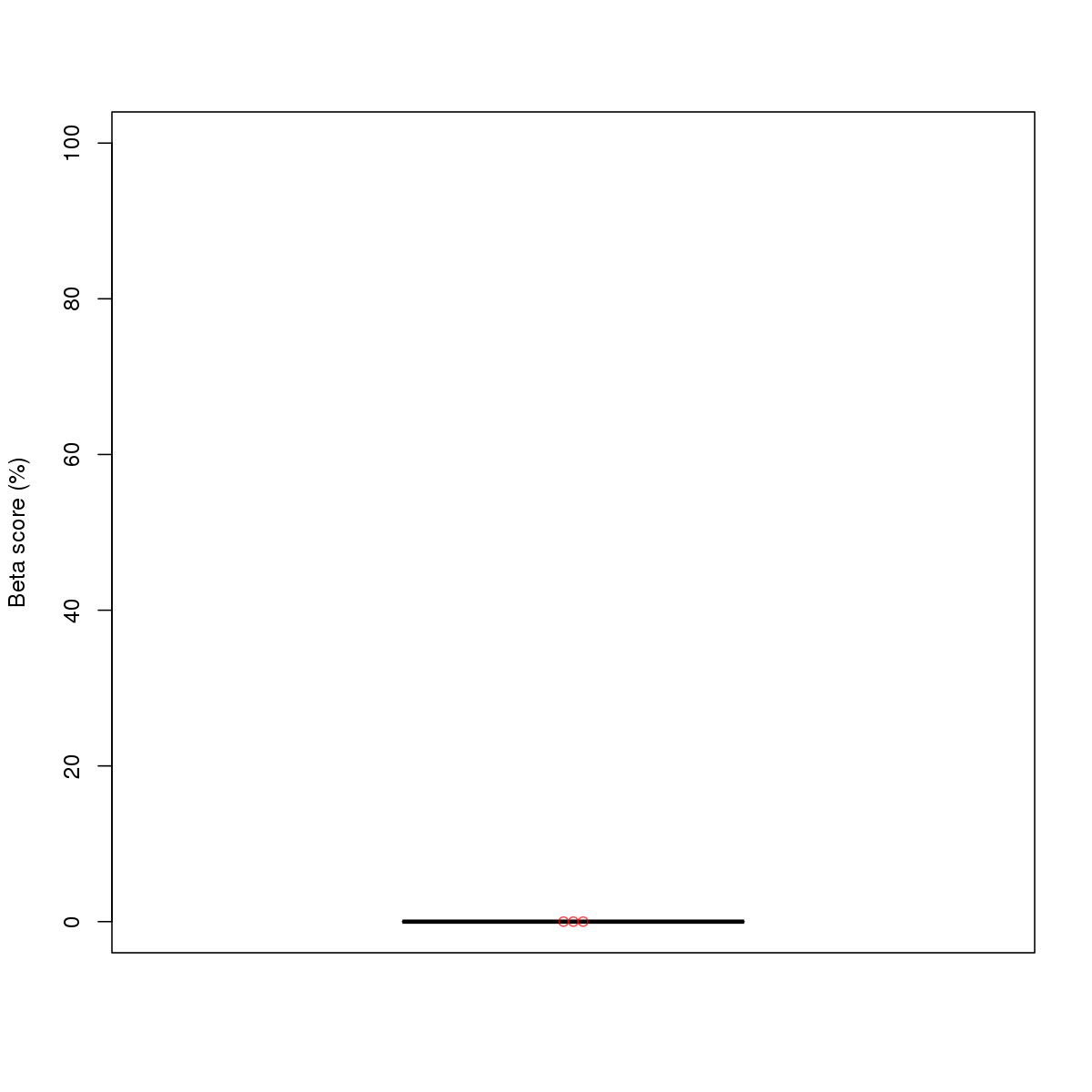 | 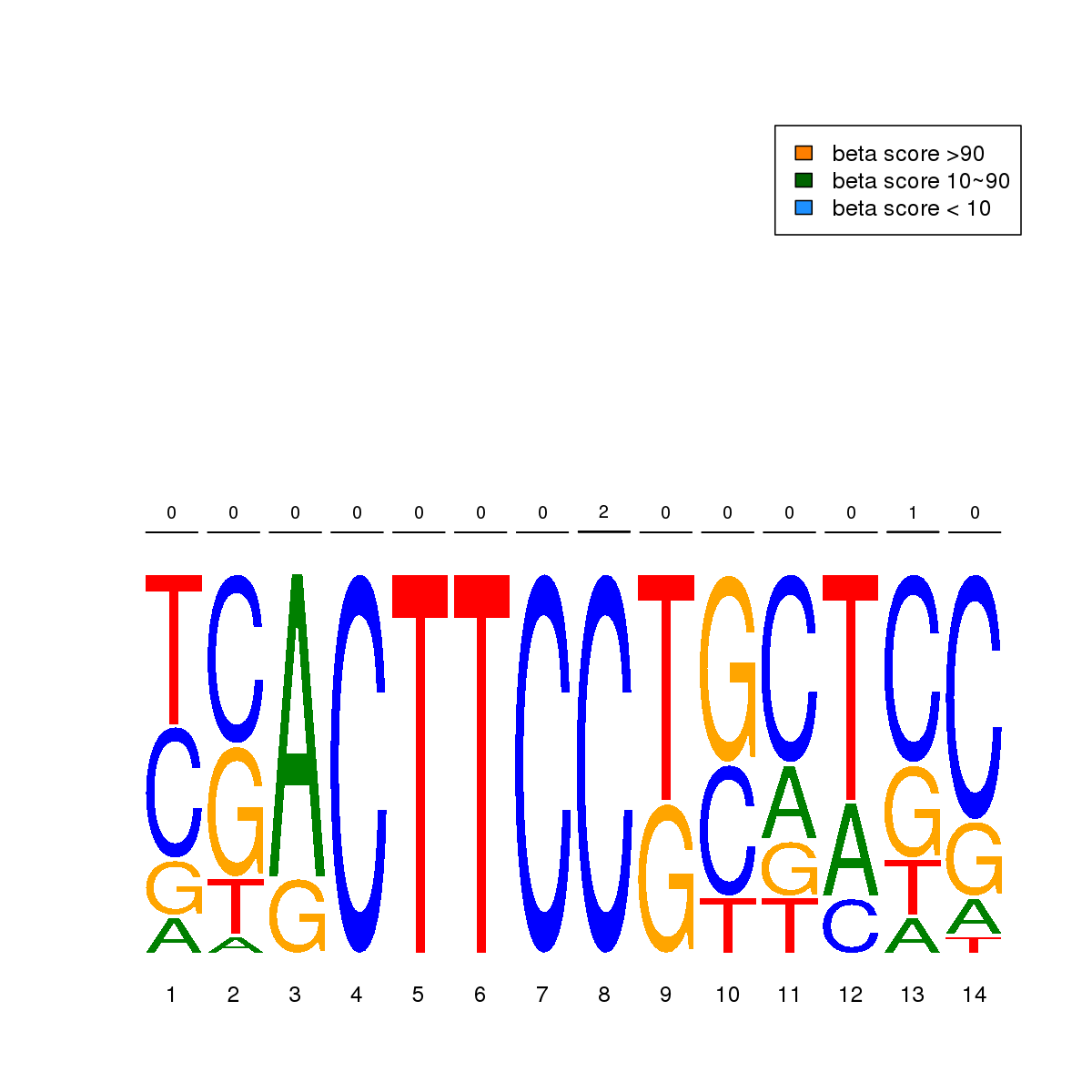 | Regions with ELF1 motif: CG methScore in motif: Beta score matrix: |
| 11 | MEL | MAZ (Details) | Factors with multiple dispersed zinc fingers | 20 | 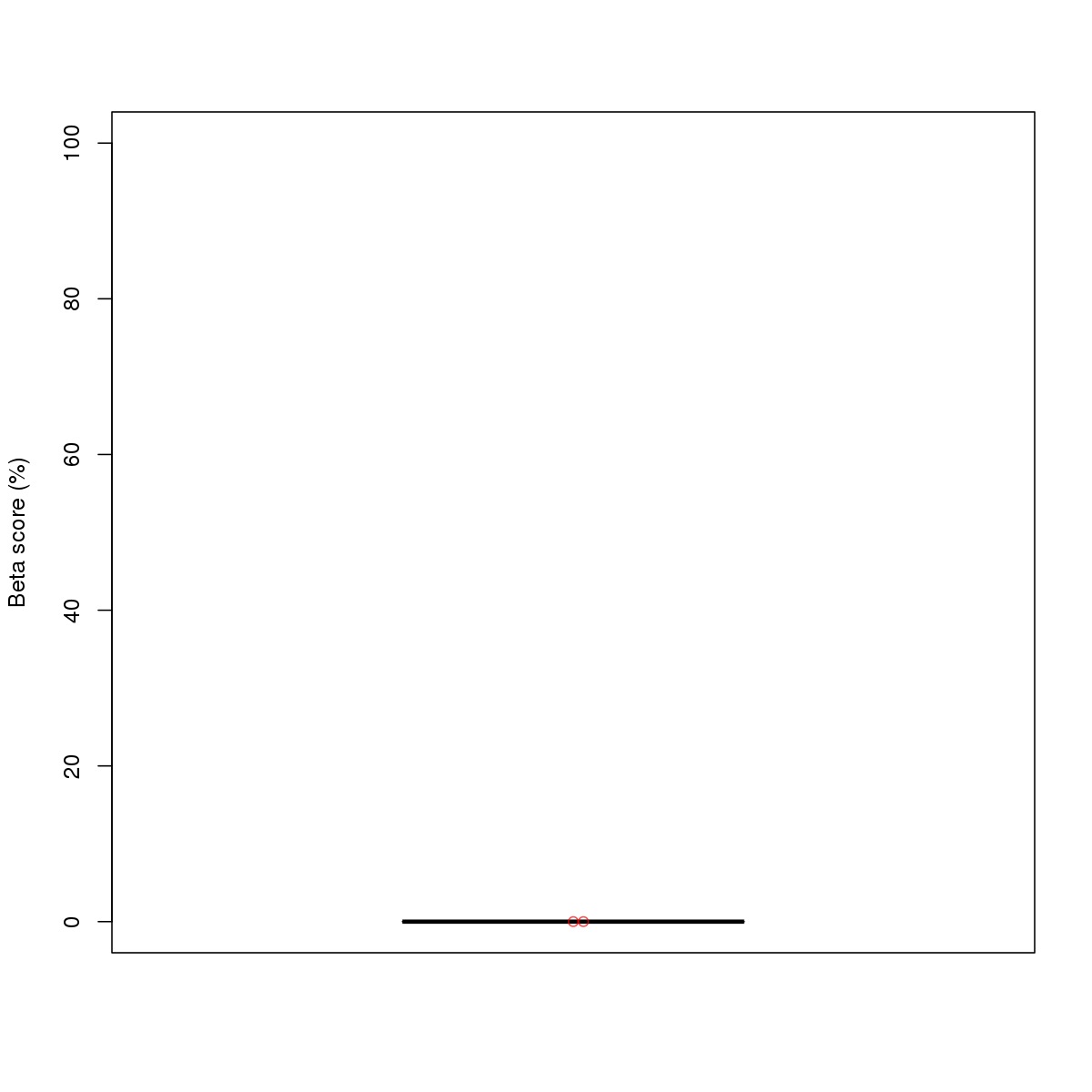 | 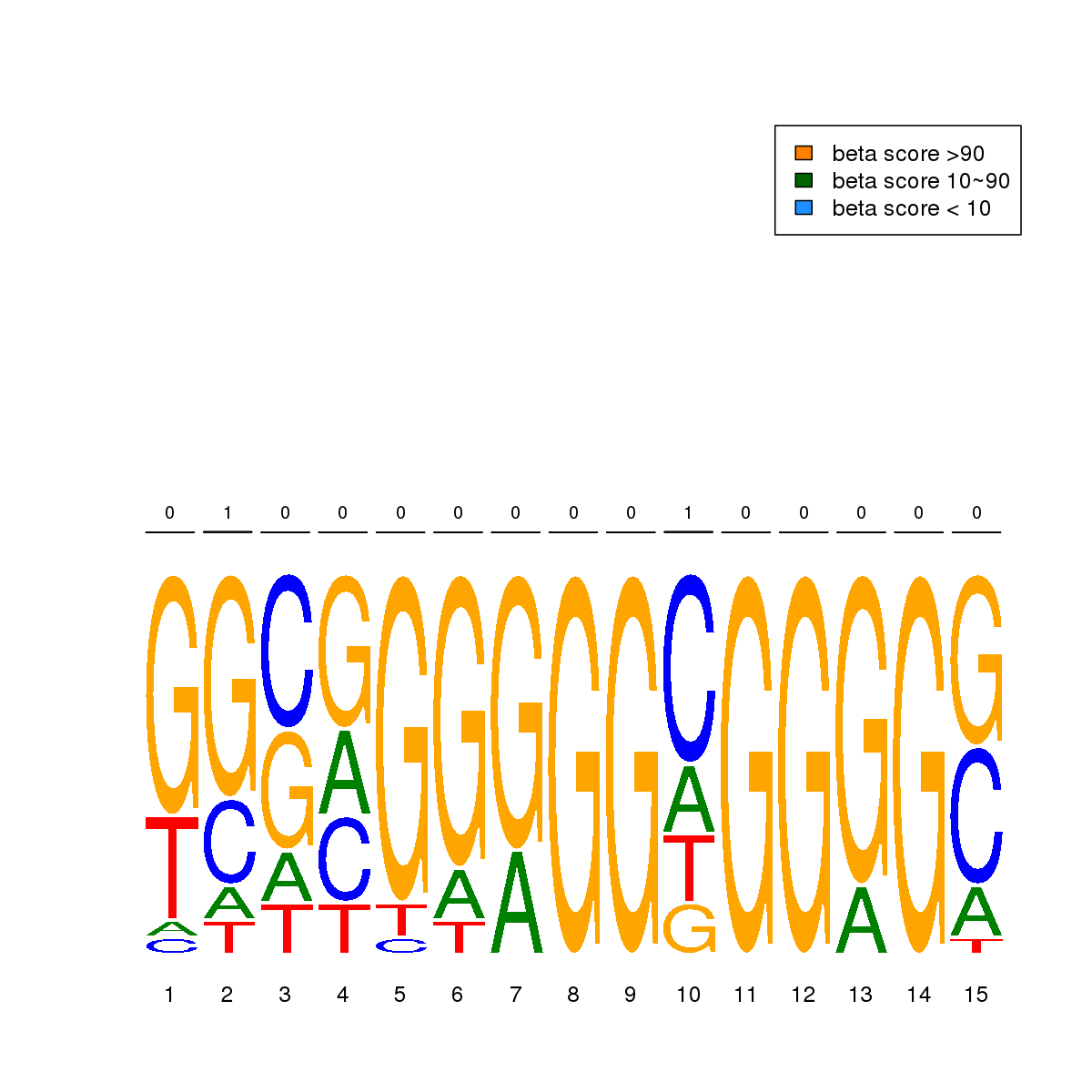 | Regions with MAZ motif: CG methScore in motif: Beta score matrix: |
| 12 | MEL | TCF12 (Details) | E2A-related factors | 13 | No CGs | 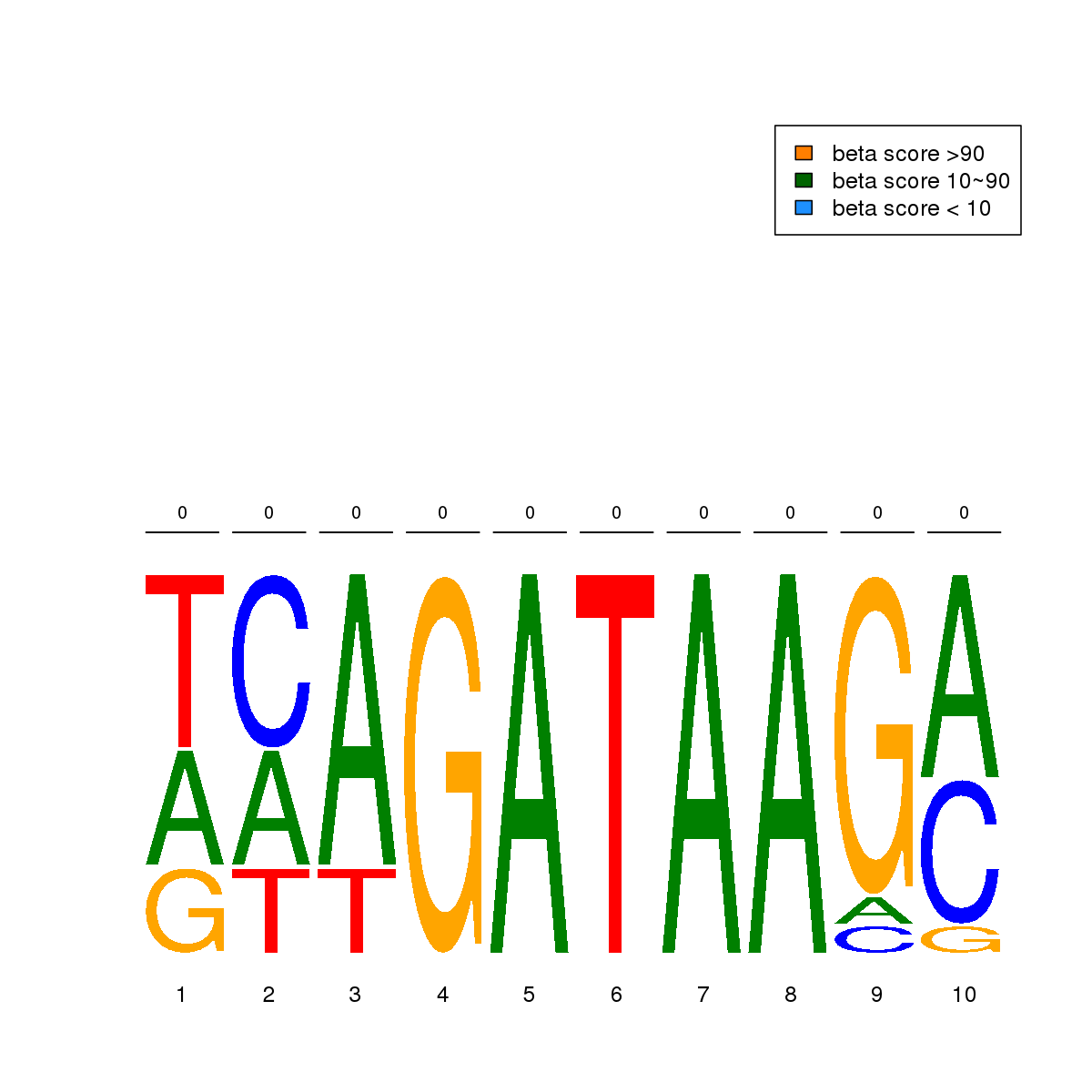 | Regions with TCF12 motif: CG methScore in motif: Beta score matrix: |
| 13 | MEL | CHD2 (Details) | Unspecified | 11 | 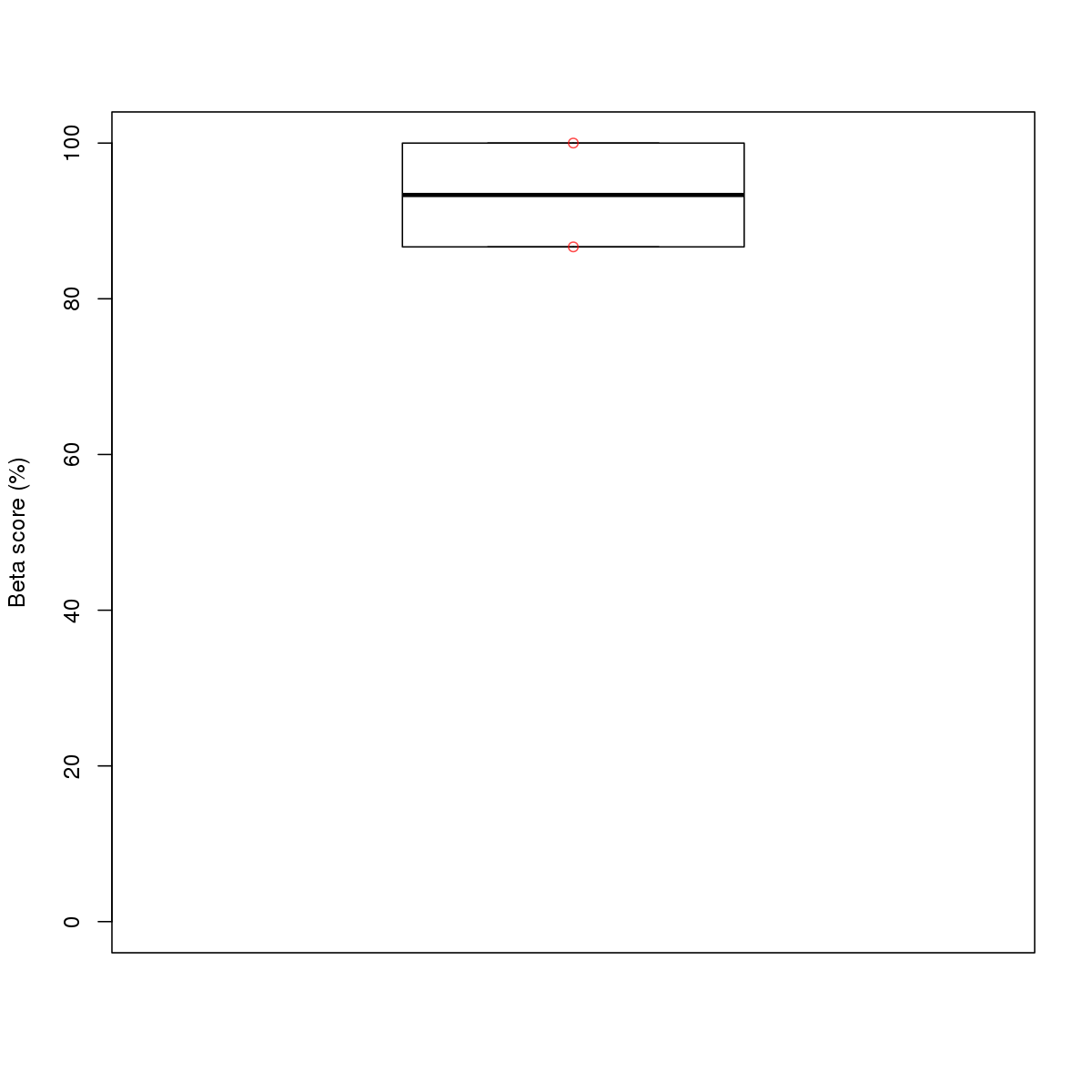 | 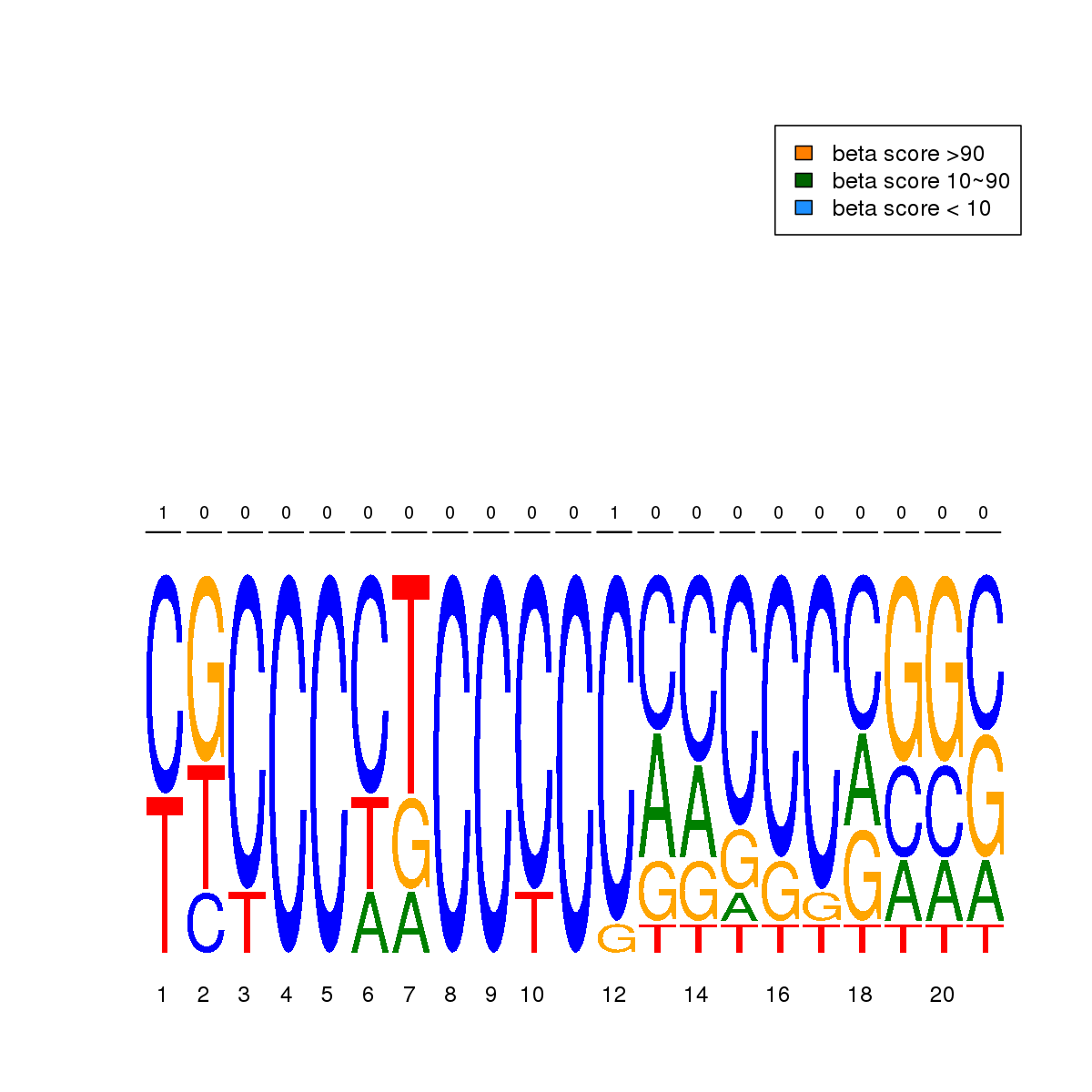 | Regions with CHD2 motif: CG methScore in motif: Beta score matrix: |
| 14 | MEL | ETS1 (Details) | Ets-related factors | 10 | No CGs | 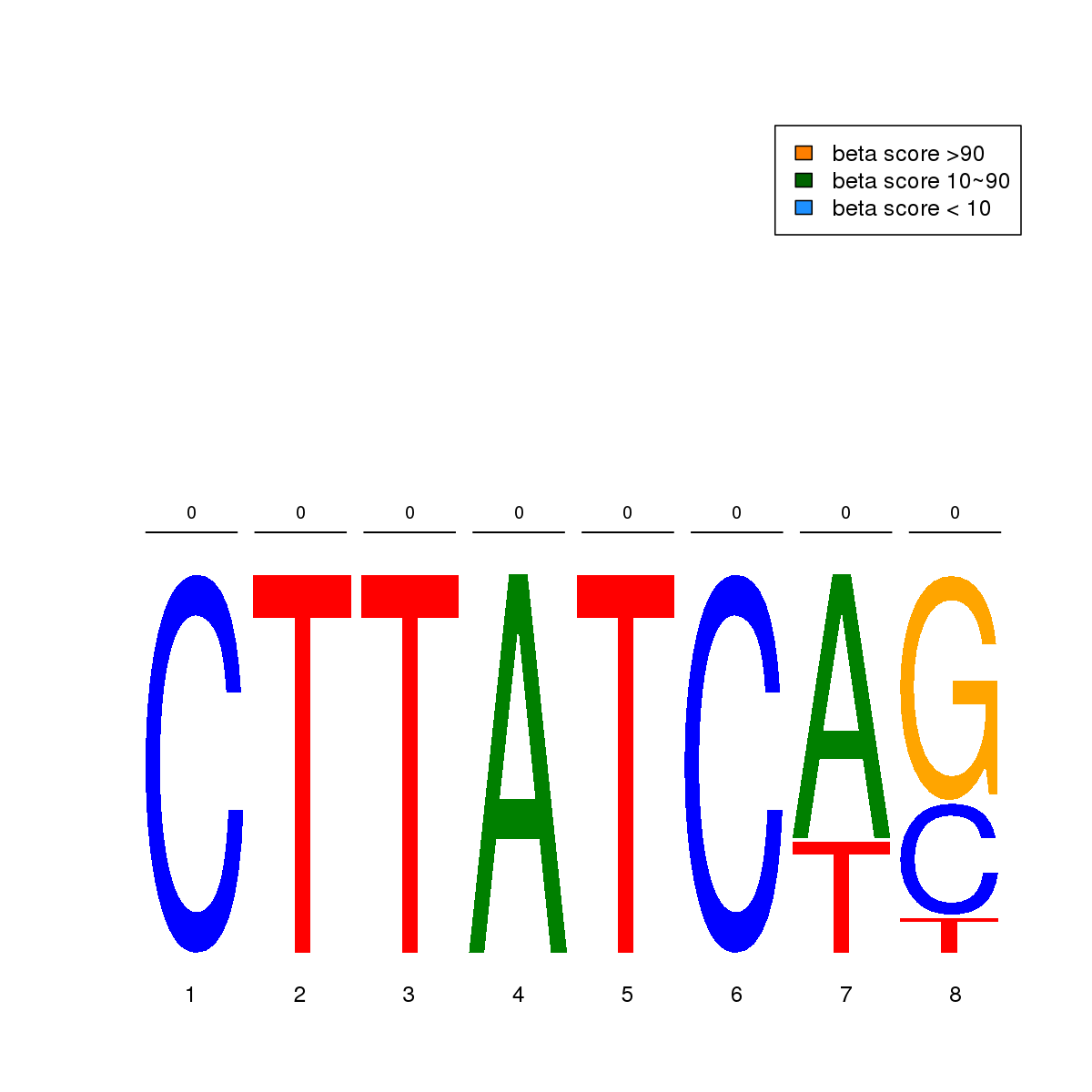 | Regions with ETS1 motif: CG methScore in motif: Beta score matrix: |
| 15 | MEL | CTCF (Details) | More than 3 adjacent zinc finger factors | 9 |  | 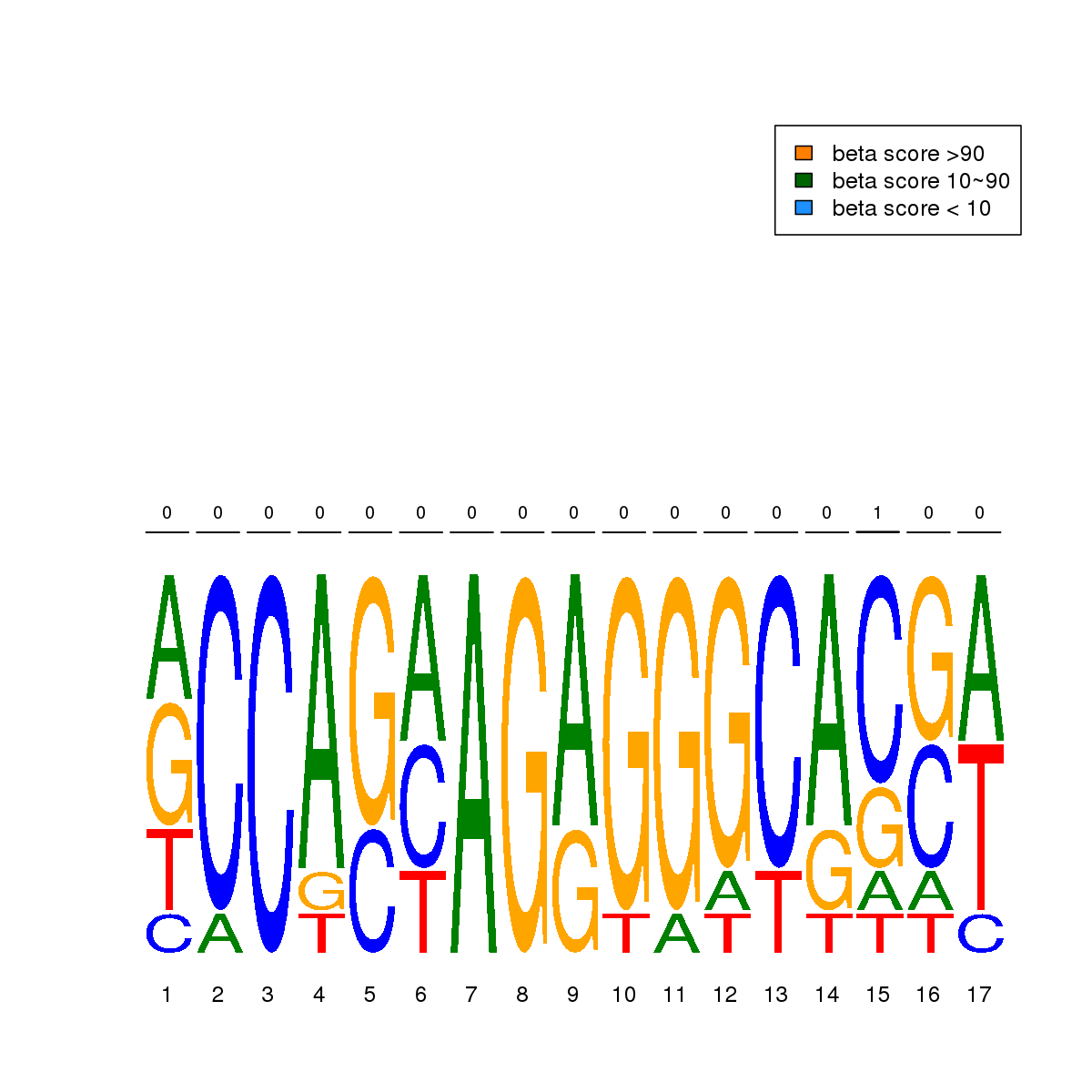 | Regions with CTCF motif: CG methScore in motif: Beta score matrix: |
| 16 | MEL | GABPA (Details) | Ets-related factors | 5 | 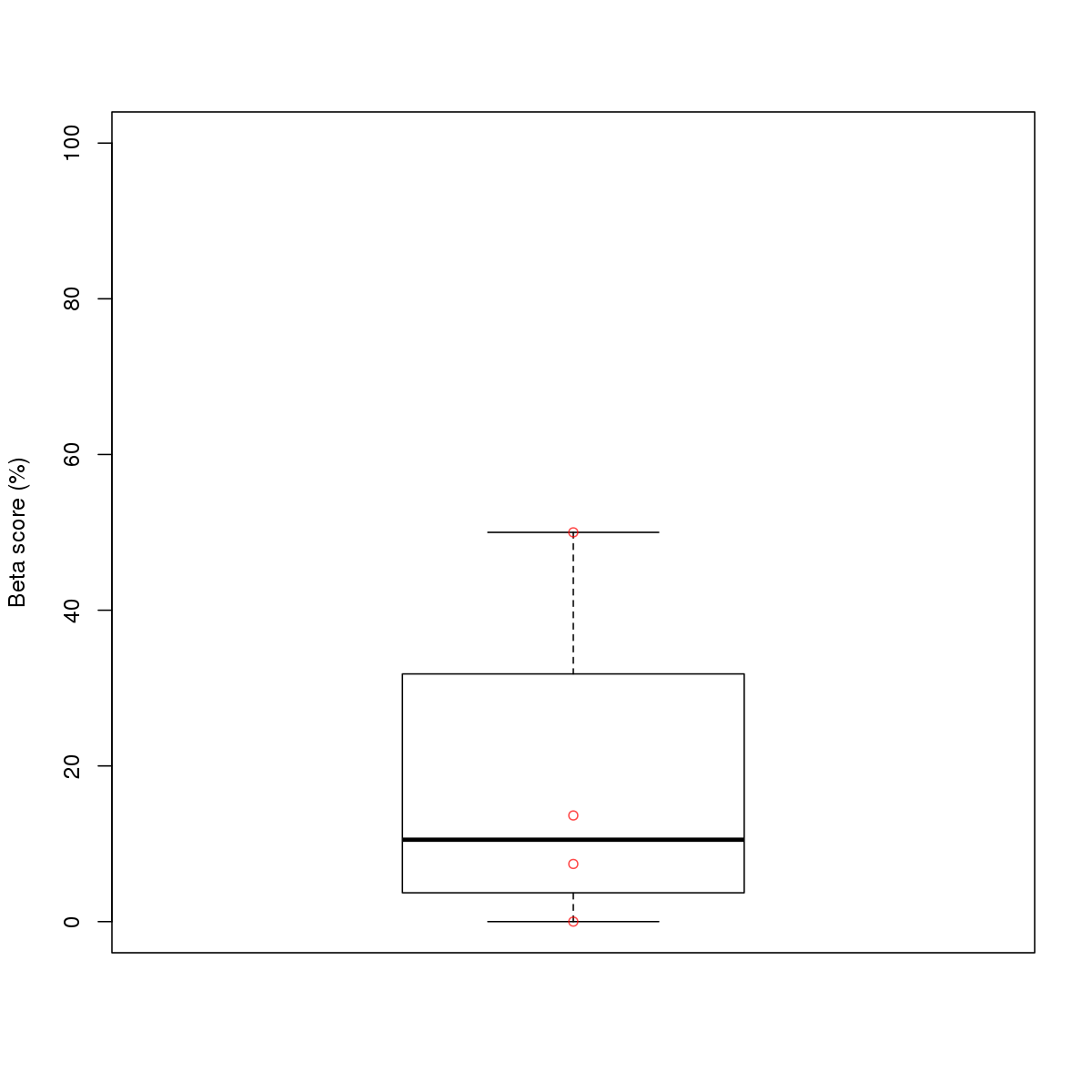 | 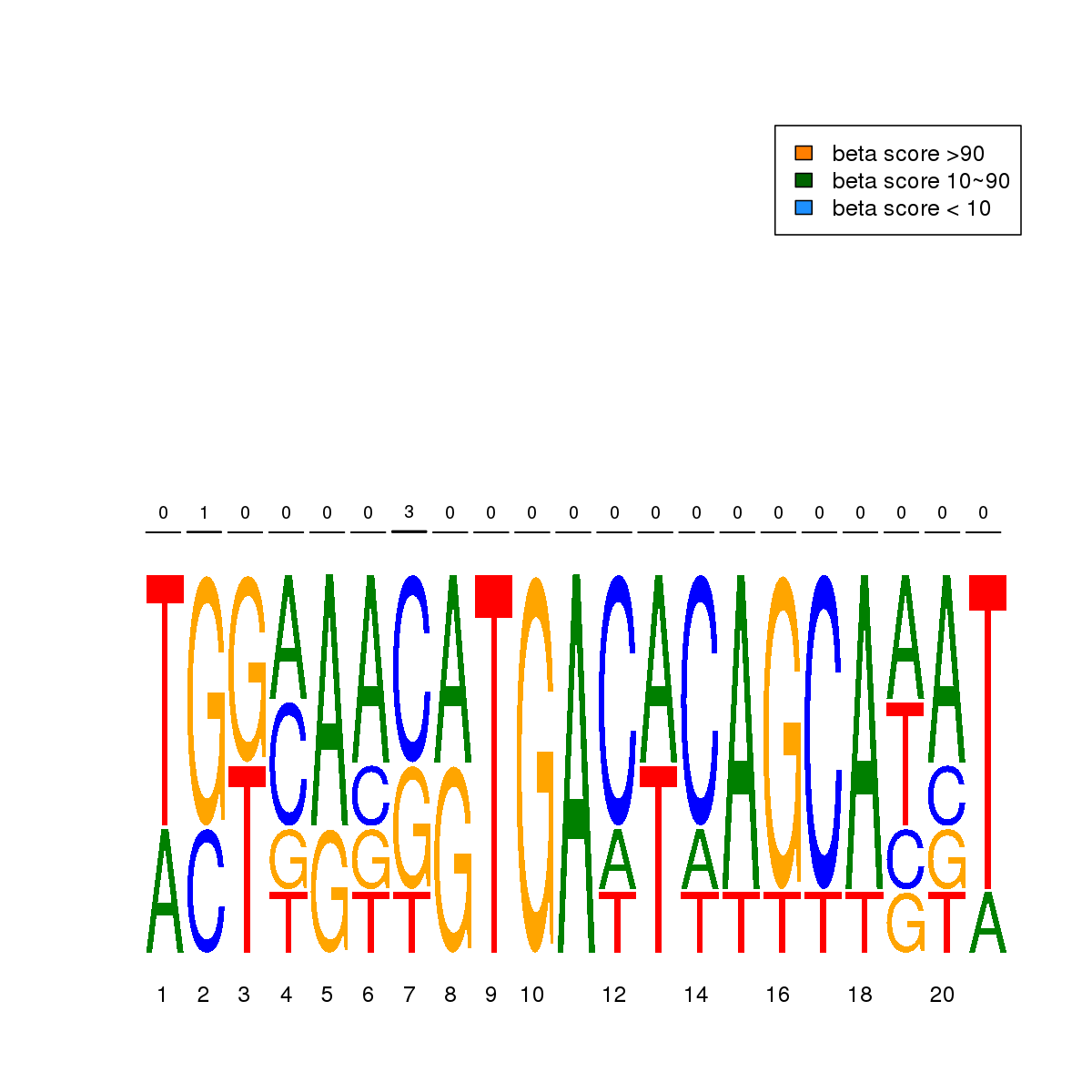 | Regions with GABPA motif: CG methScore in motif: Beta score matrix: |
| 17 | MEL | MAFK (Details) | Maf-related factors | 3 |  | 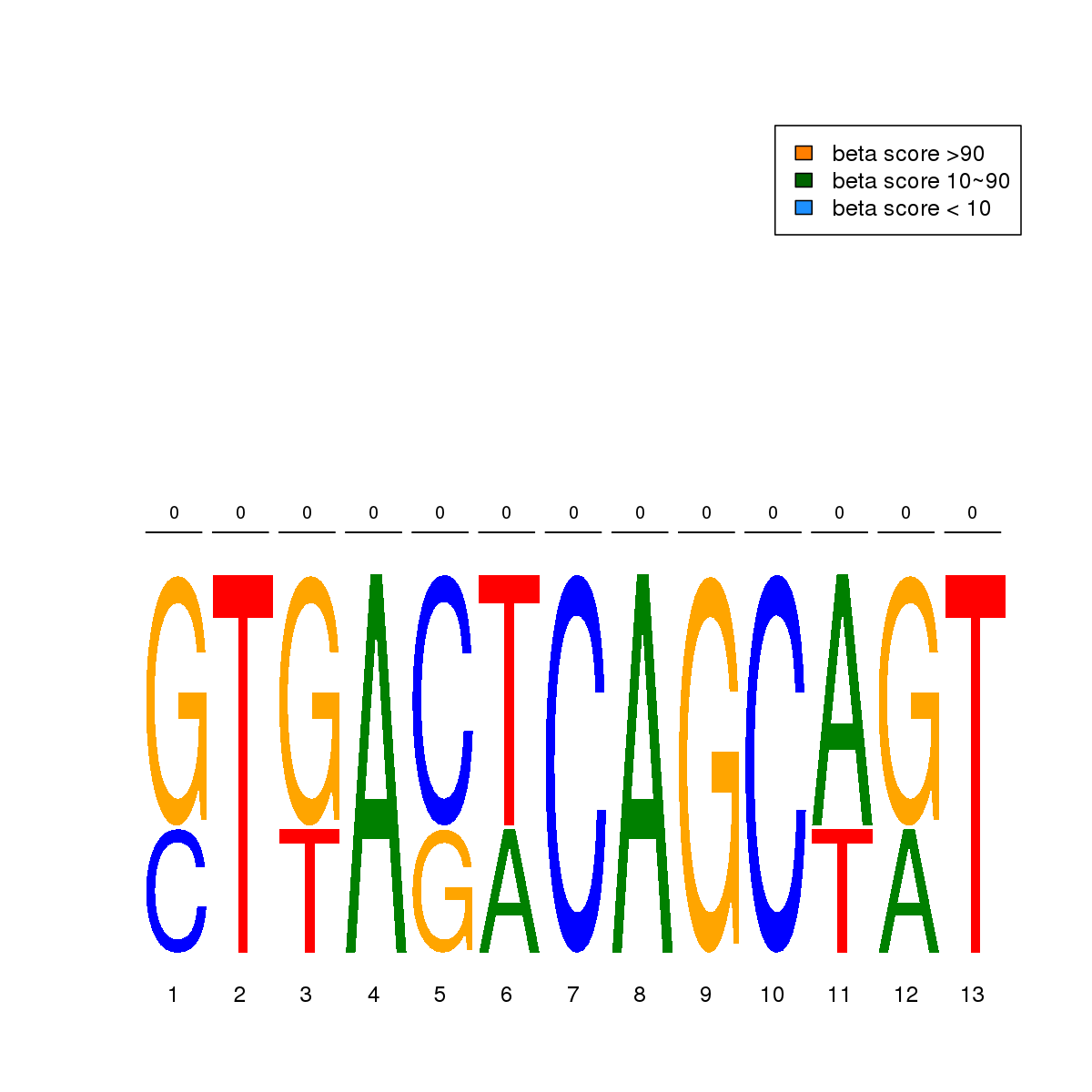 | Regions with MAFK motif: CG methScore in motif: Beta score matrix: |
| 18 | MEL | MYB (Details) | Myb/SANT domain factors | 3 | No CGs | 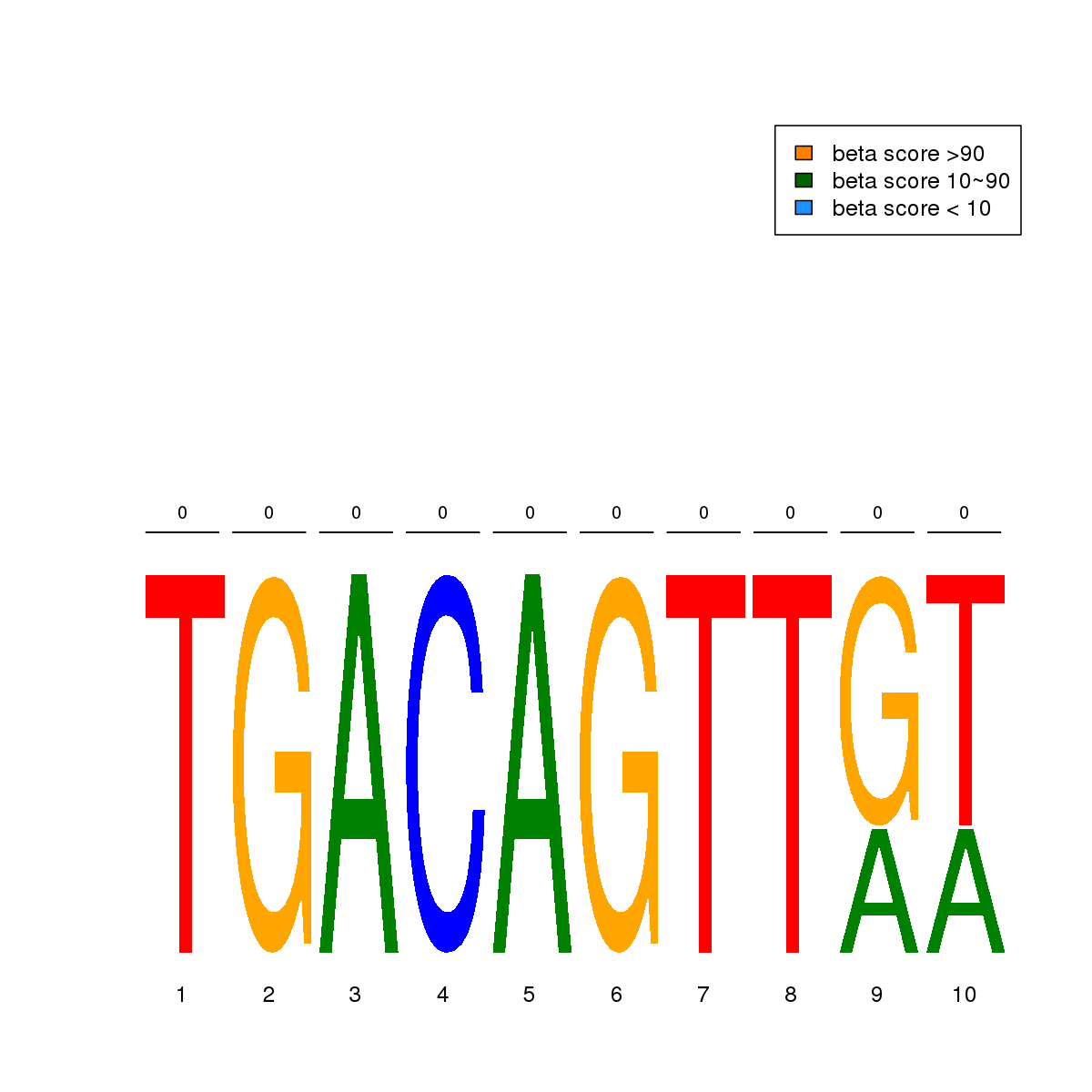 | Regions with MYB motif: CG methScore in motif: Beta score matrix: |
| 19 | MEL | ZKSCAN1 (Details) | More than 3 adjacent zinc finger factors | 1 | 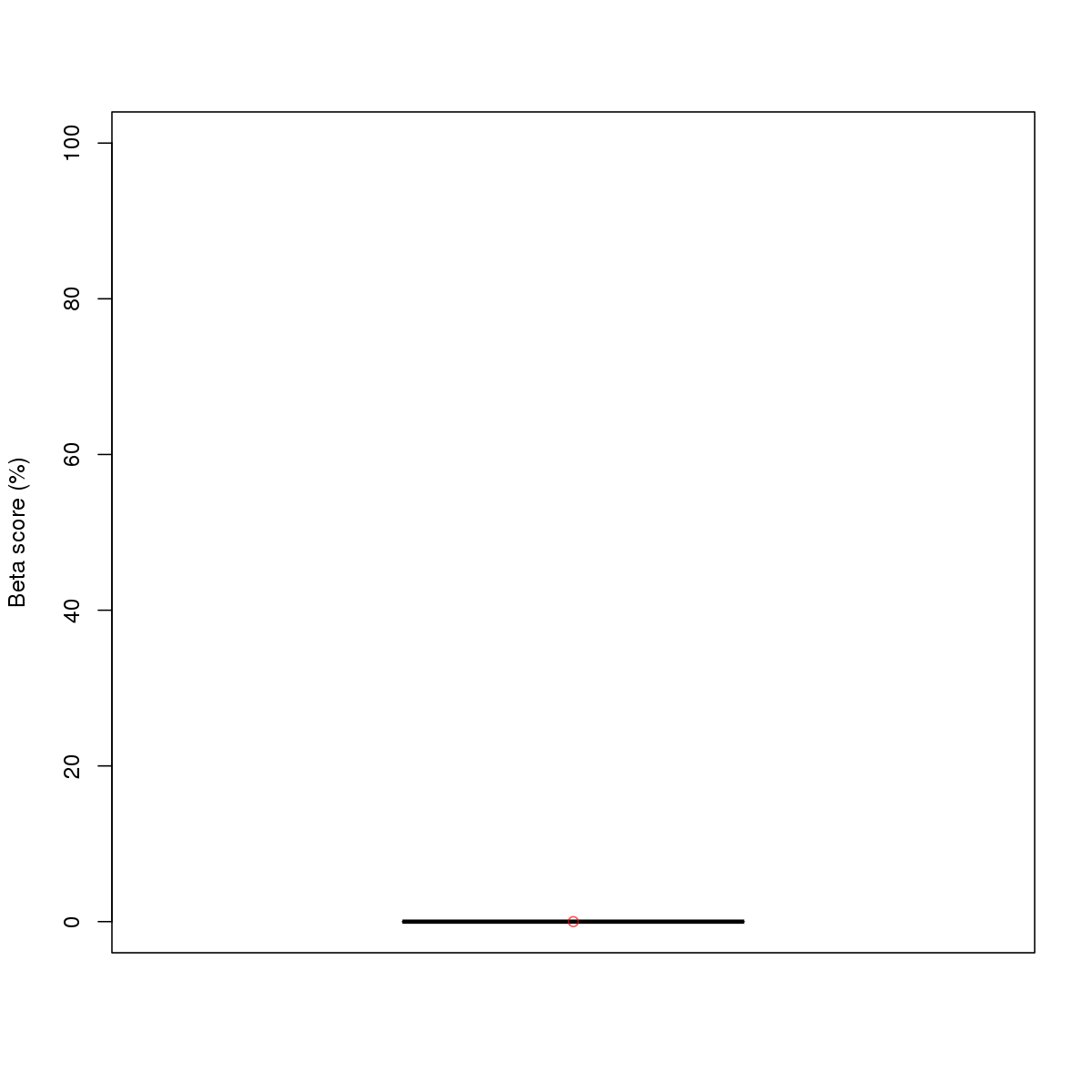 | 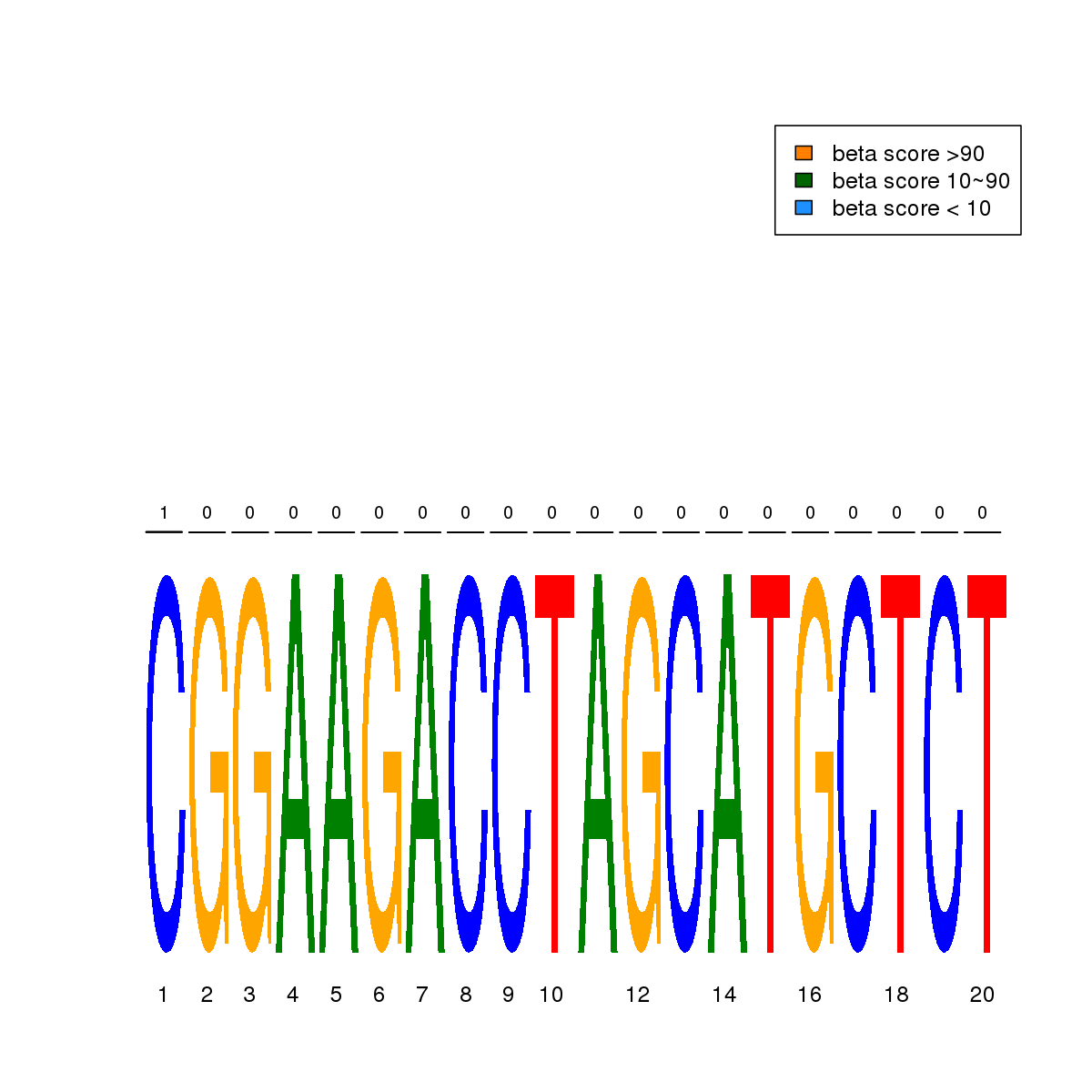 | Regions with ZKSCAN1 motif: CG methScore in motif: Beta score matrix: |