Genome assembly: hg38
Demo dataset (CEBPb ChIP-seq experiment in HCT116 [ENCSR000BSD])
| ID | Cell type | TF | Family | Number of input regions with TFBS | CG beta scores in the TFBS | MethMotif logo | Details |
|---|---|---|---|---|---|---|---|
| 1 | HCT116 | CEBPB (Details) | C/EBP-related | 4846 | 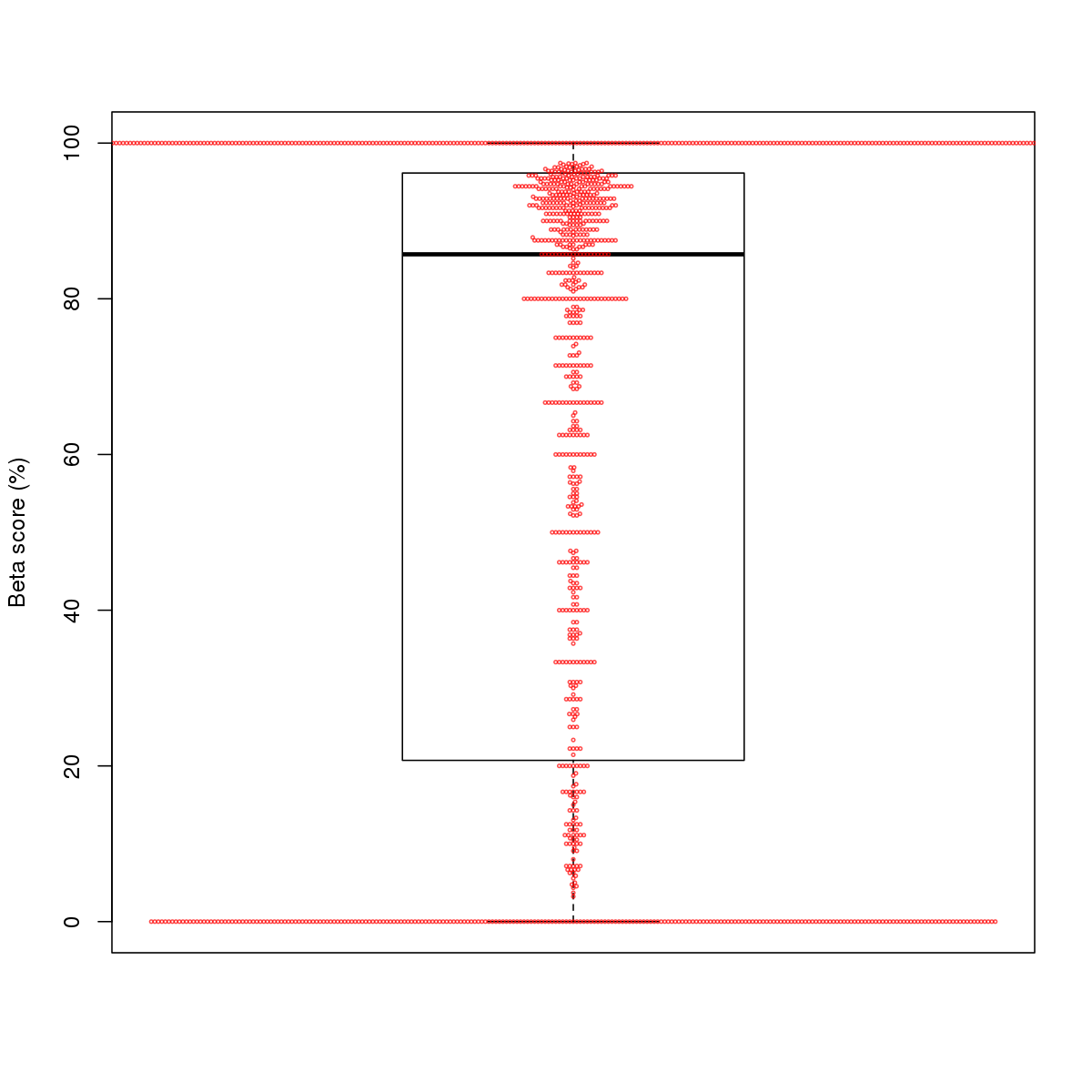 | 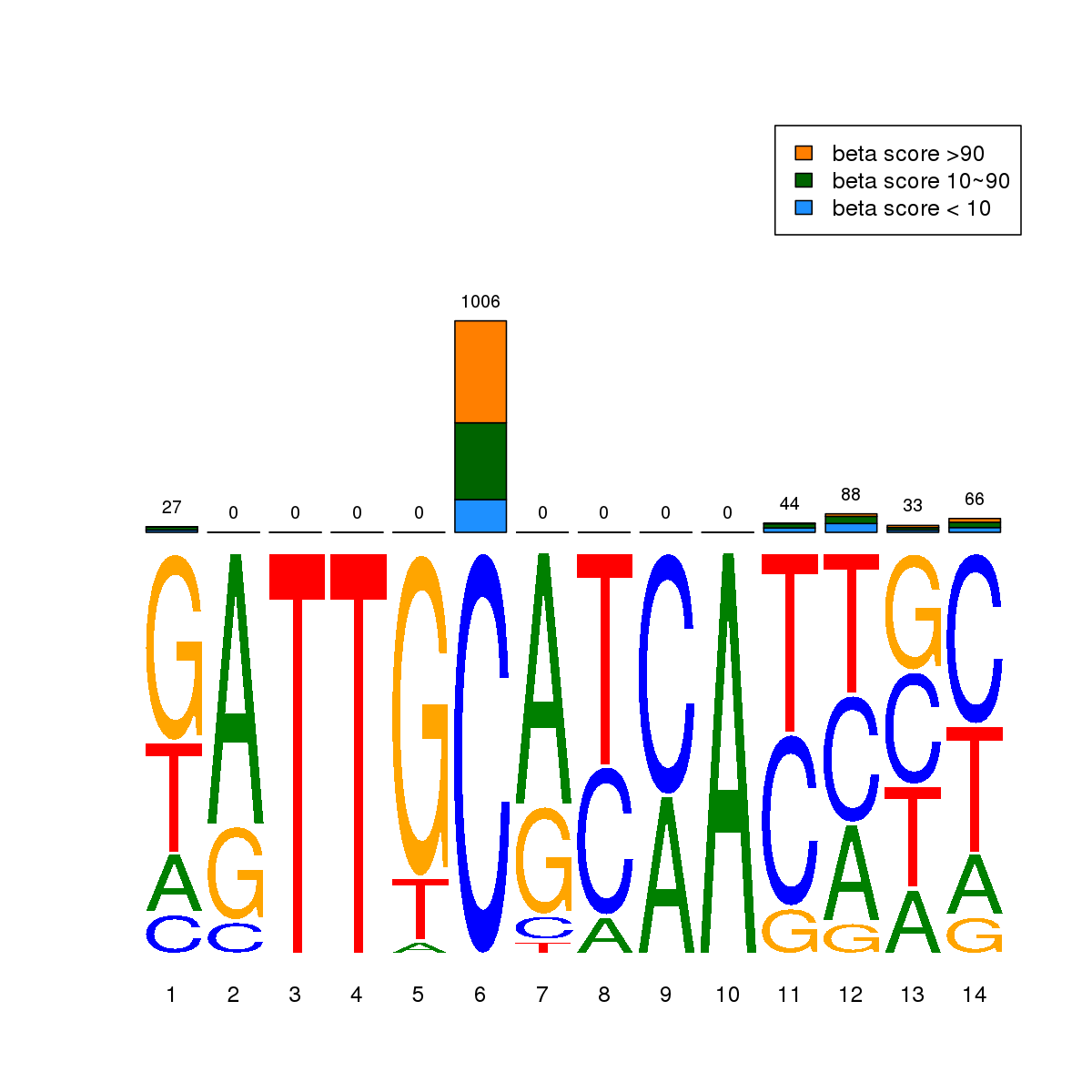 | Regions with CEBPB motif: CG methScore in motif: Beta score matrix: |
| 2 | HCT116 | ATF3 (Details) | Fos-related factors | 300 | 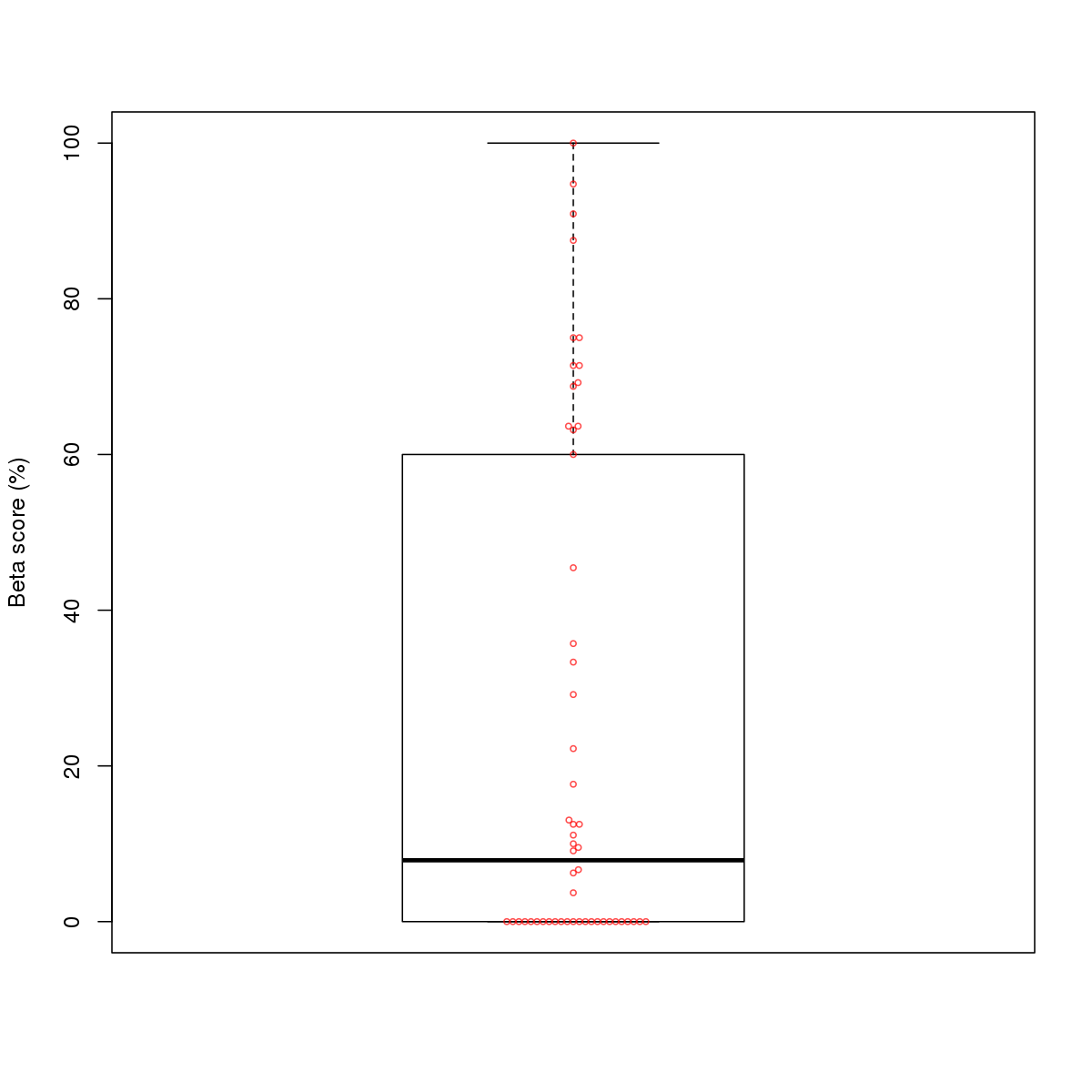 | 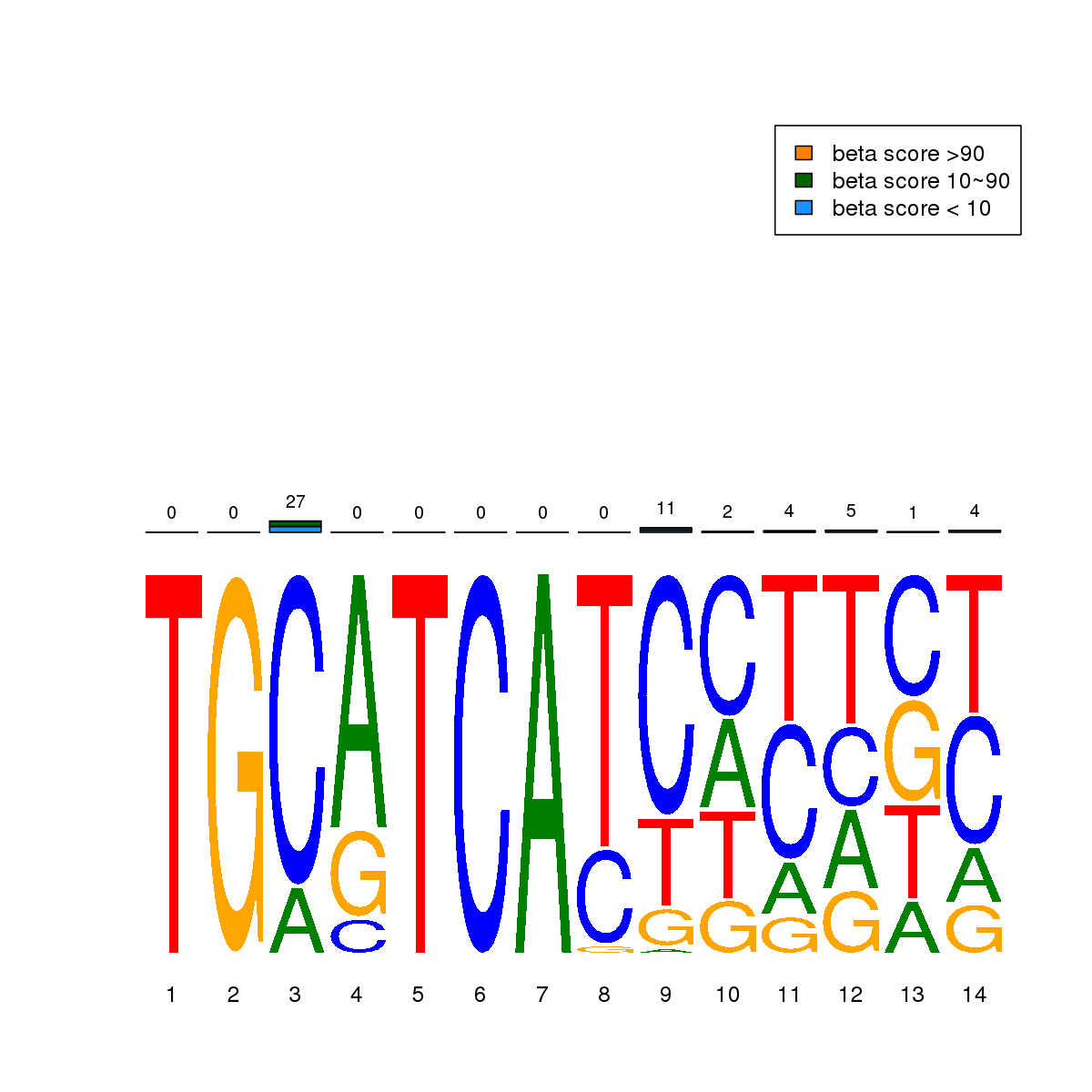 | Regions with ATF3 motif: CG methScore in motif: Beta score matrix: |
| 3 | HCT116 | TEAD4 (Details) | TEF-1-related factors | 106 | 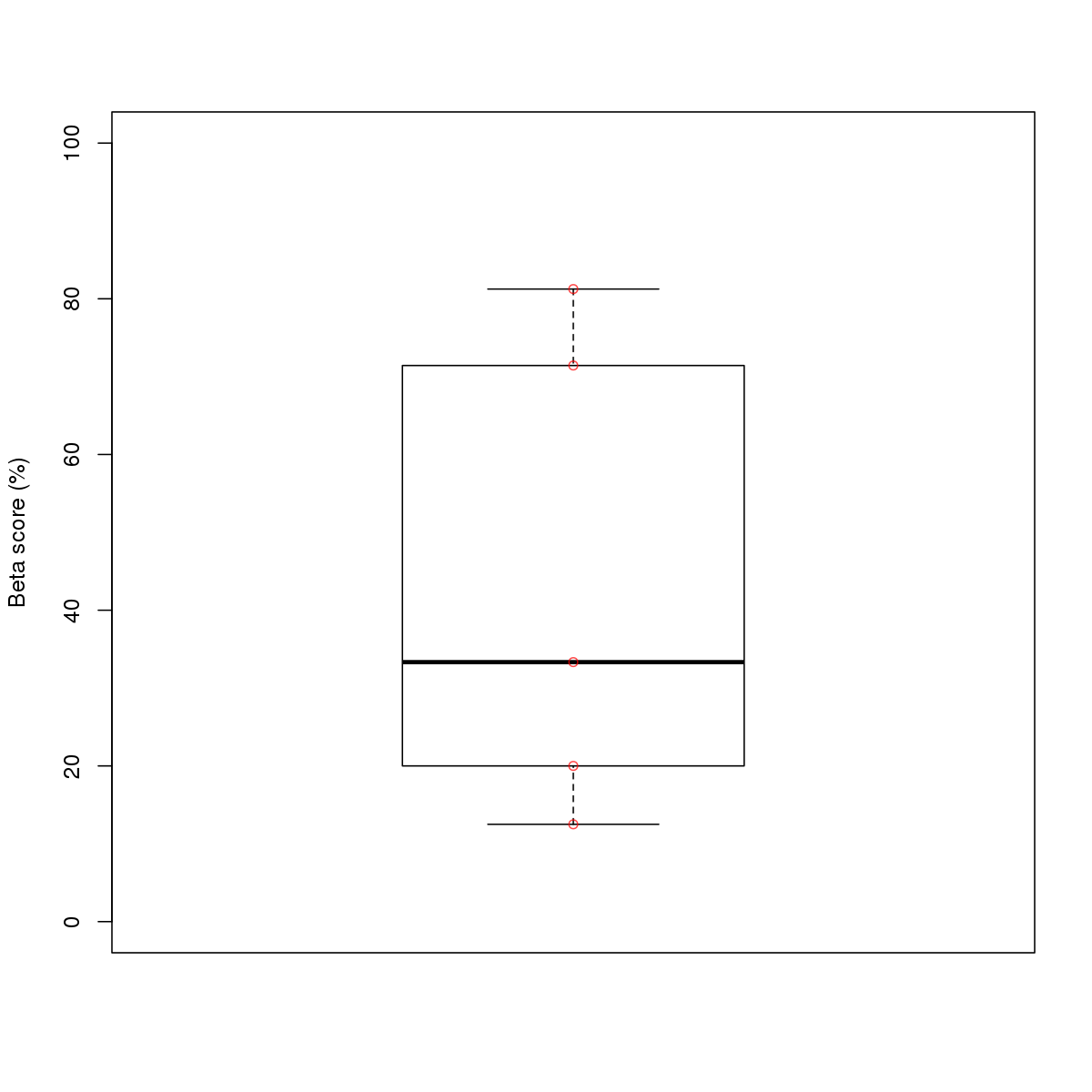 |  | Regions with TEAD4 motif: CG methScore in motif: Beta score matrix: |
| 4 | HCT116 | FOSL1 (Details) | Fos-related factors | 91 | 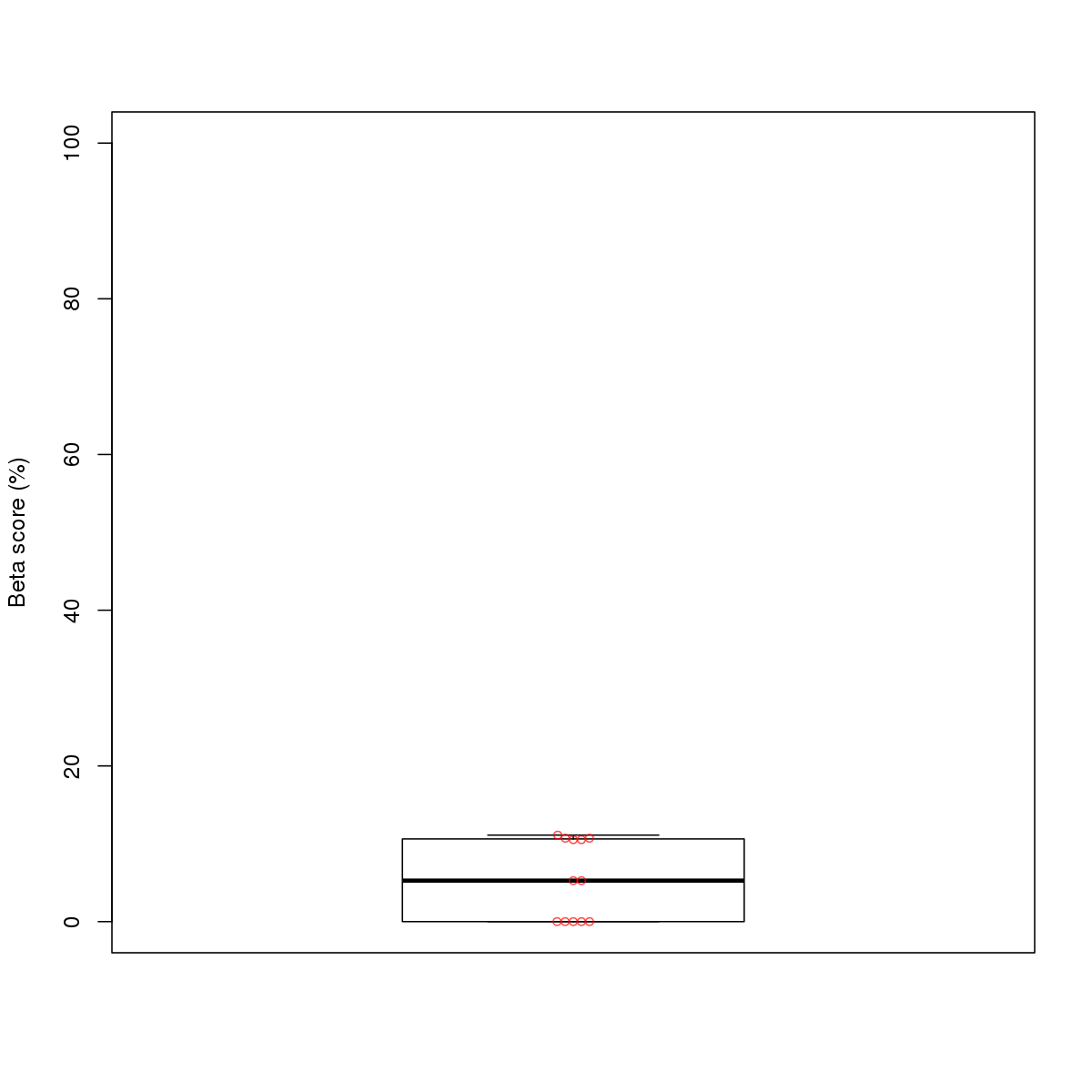 | 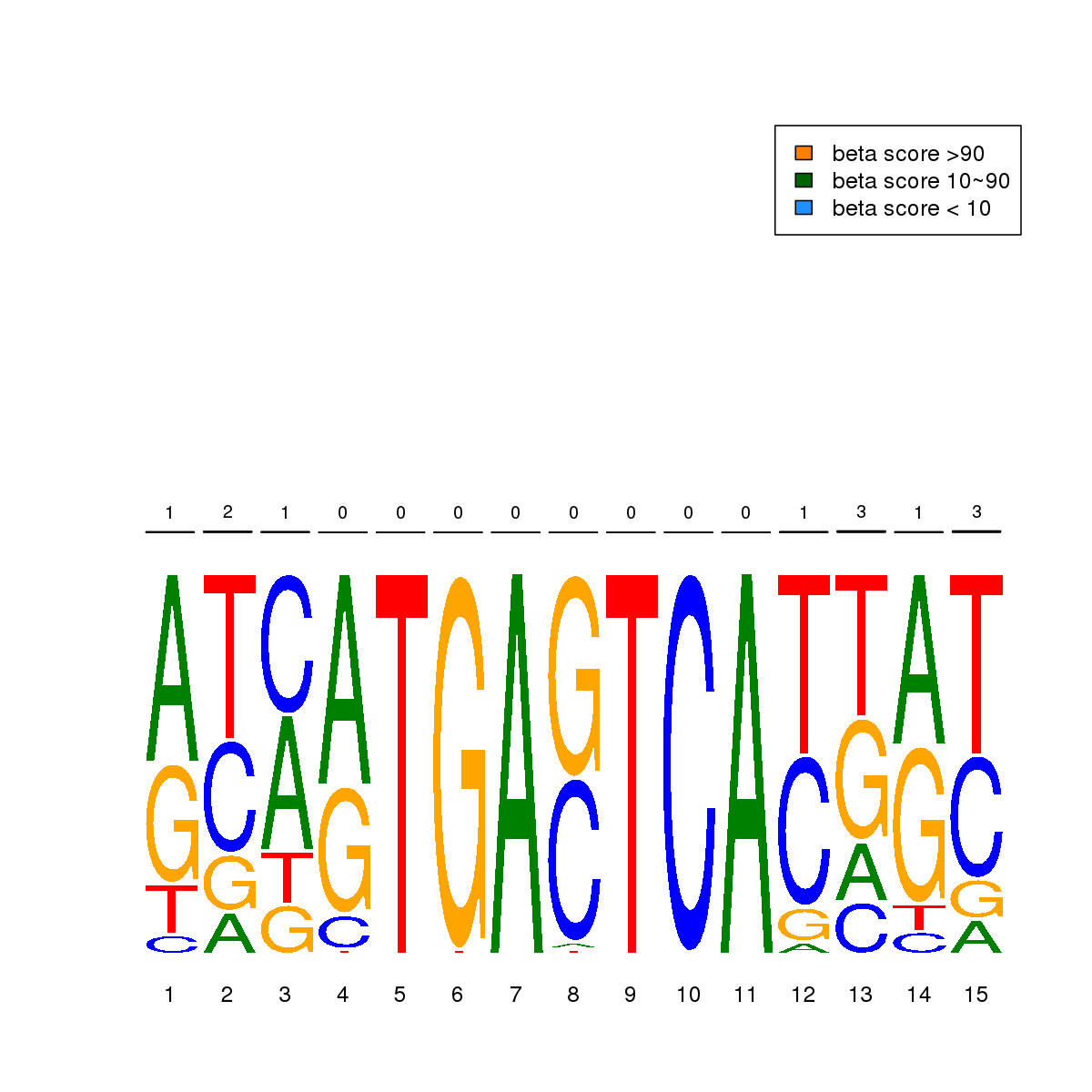 | Regions with FOSL1 motif: CG methScore in motif: Beta score matrix: |
| 5 | HCT116 | JUND (Details) | Jun-related factors | 74 | 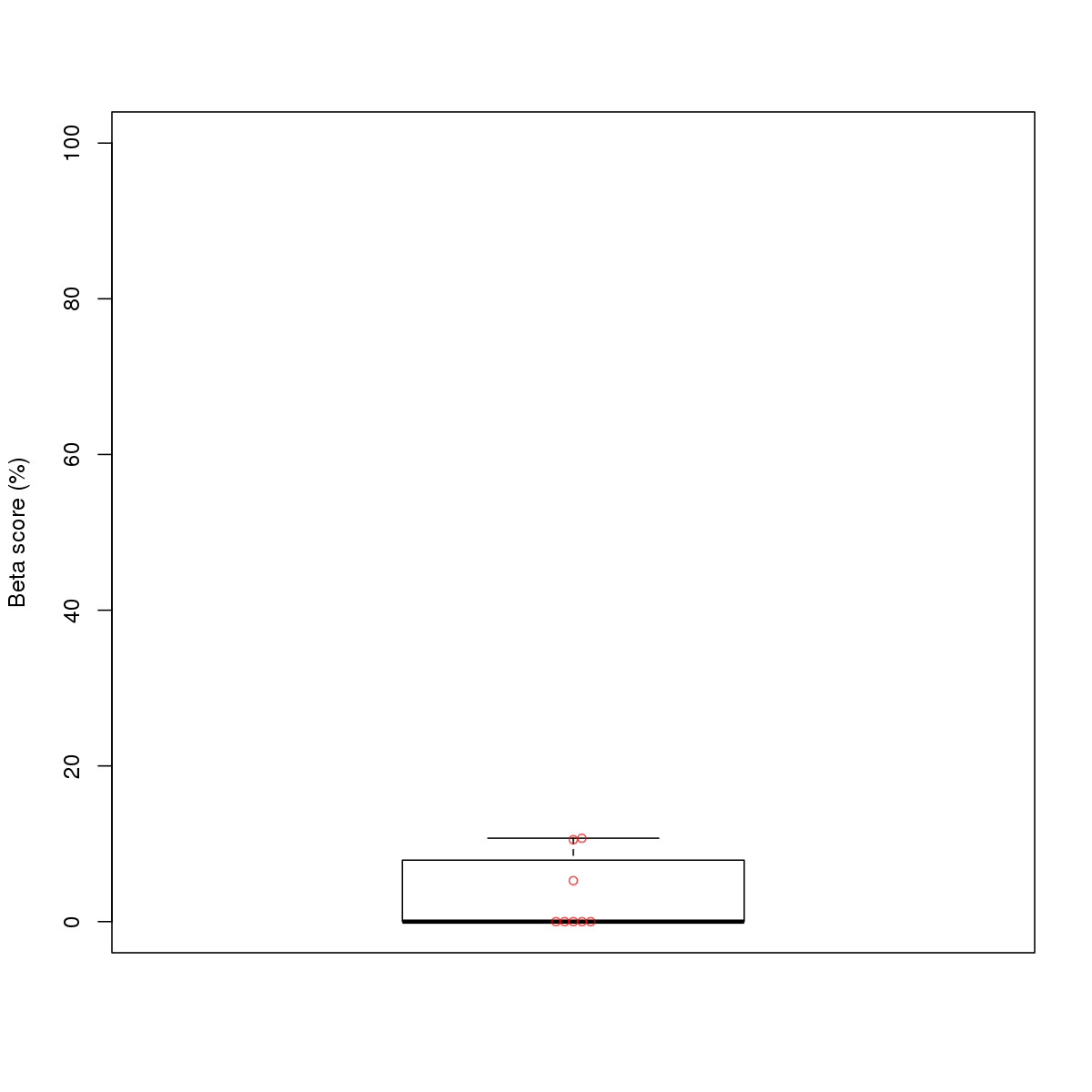 |  | Regions with JUND motif: CG methScore in motif: Beta score matrix: |
| 6 | HCT116 | ELF1 (Details) | Ets-related factors | 17 | 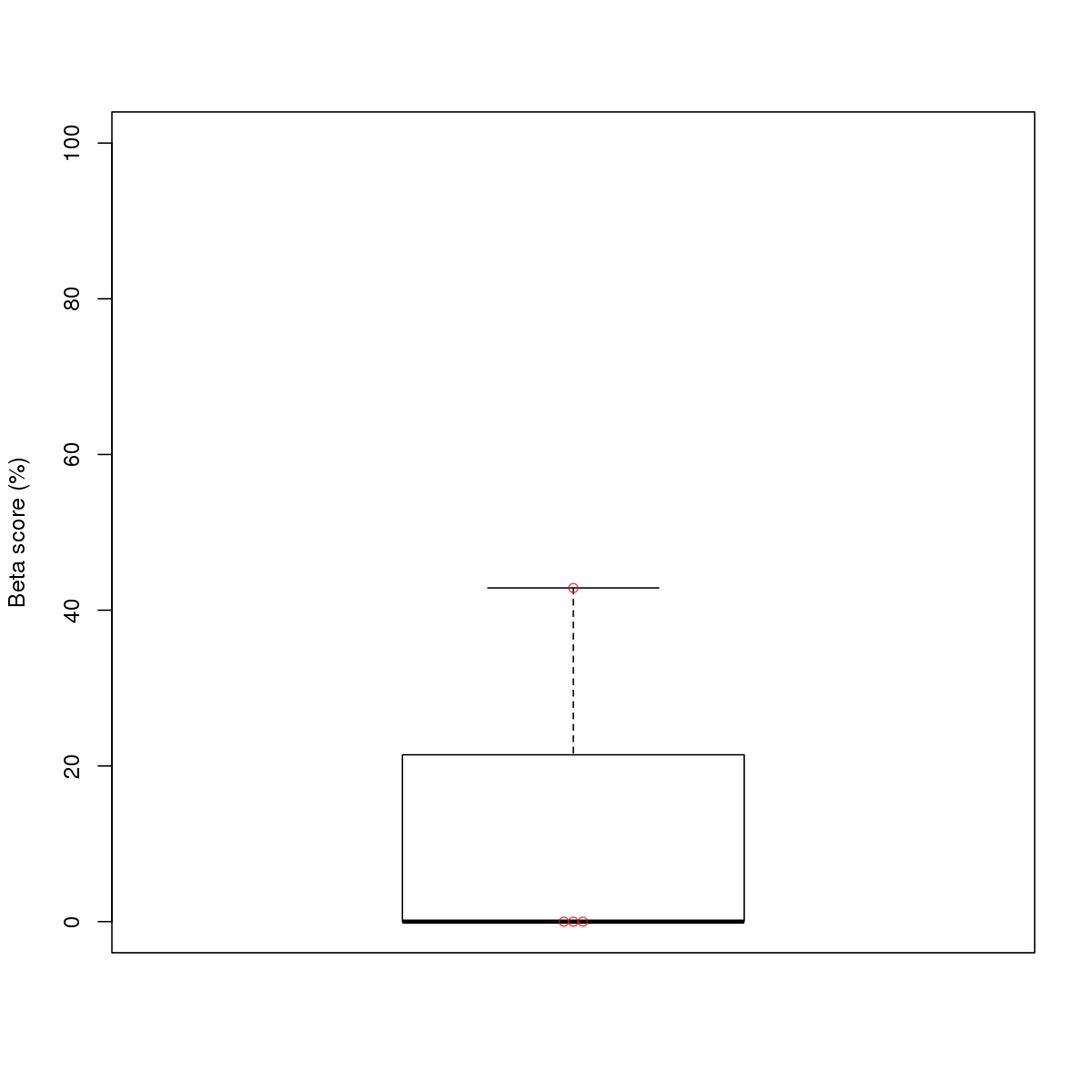 |  | Regions with ELF1 motif: CG methScore in motif: Beta score matrix: |
| 7 | HCT116 | CTCF (Details) | More than 3 adjacent zinc finger factors | 10 | 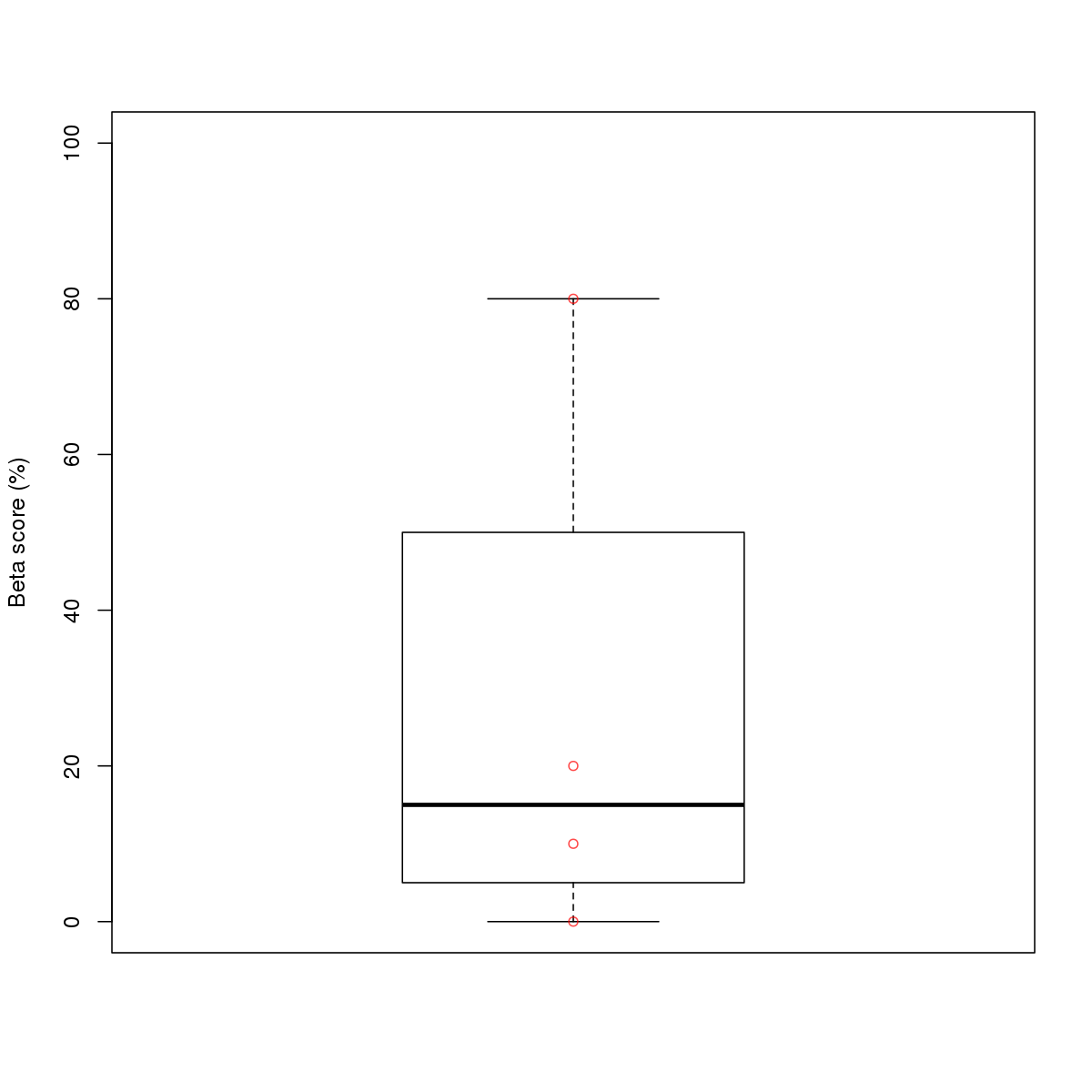 | 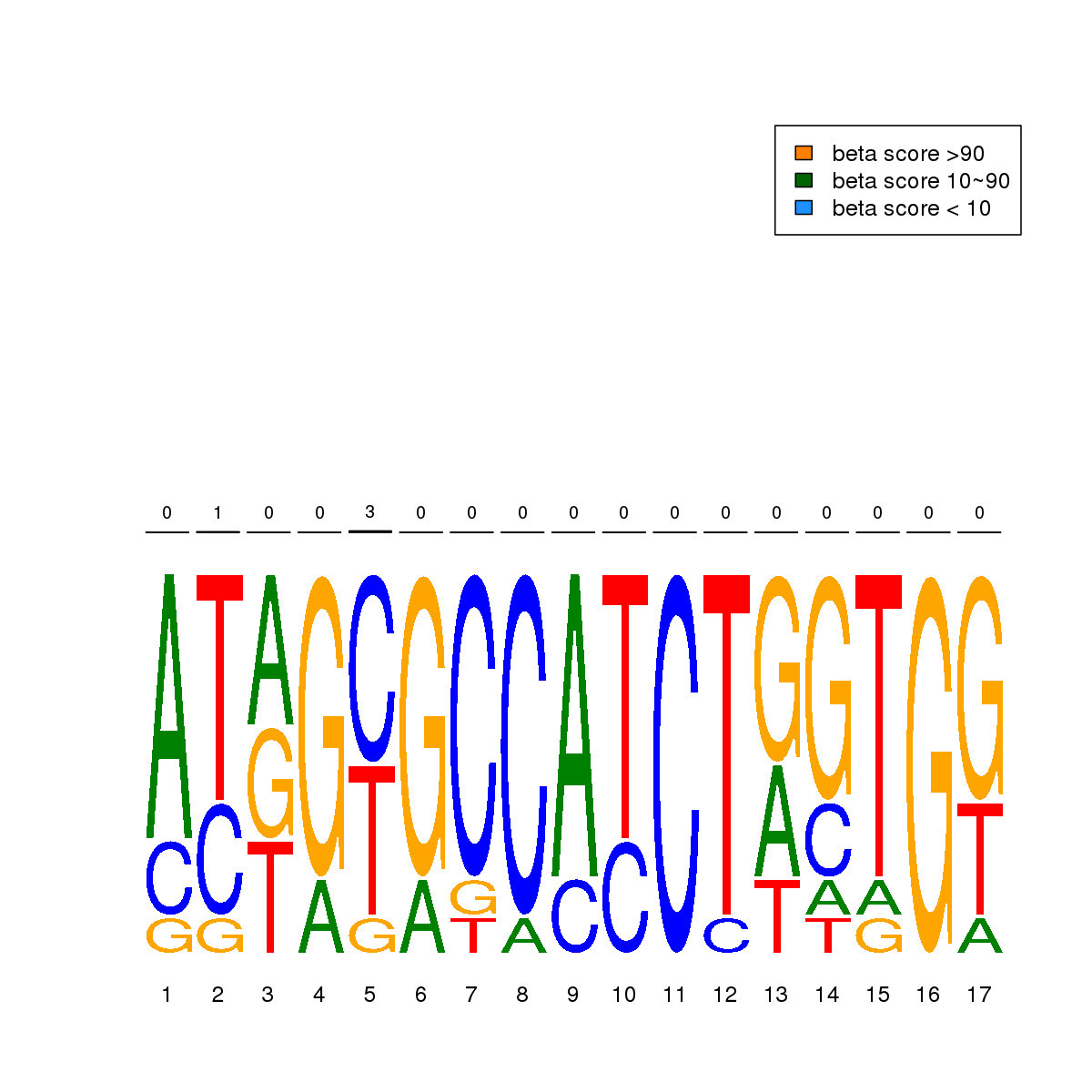 | Regions with CTCF motif: CG methScore in motif: Beta score matrix: |
| 8 | HCT116 | TCF7L2 (Details) | TCF-7-related factors | 10 | 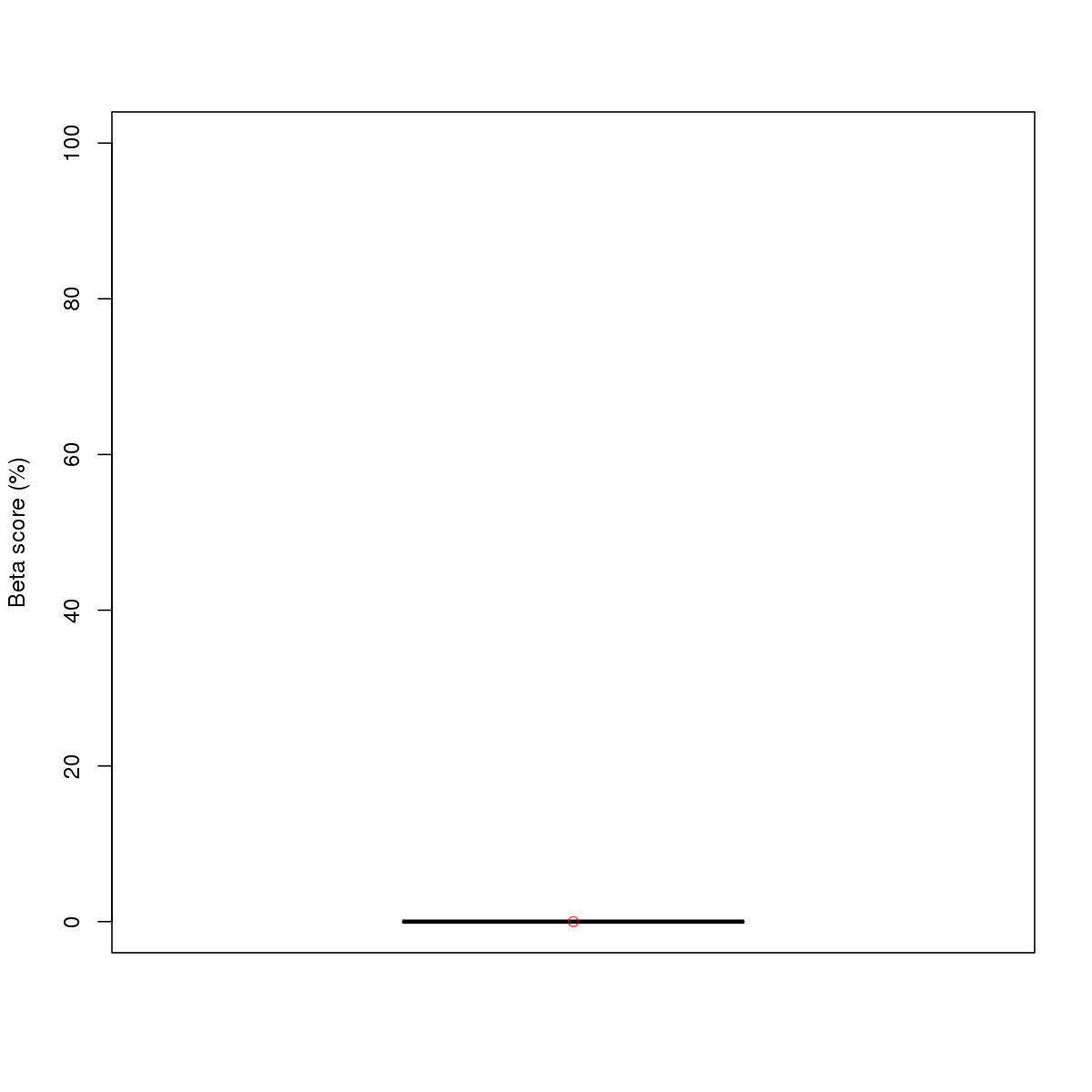 |  | Regions with TCF7L2 motif: CG methScore in motif: Beta score matrix: |
| 9 | HCT116 | USF1 (Details) | bHLH-ZIP factors | 7 | 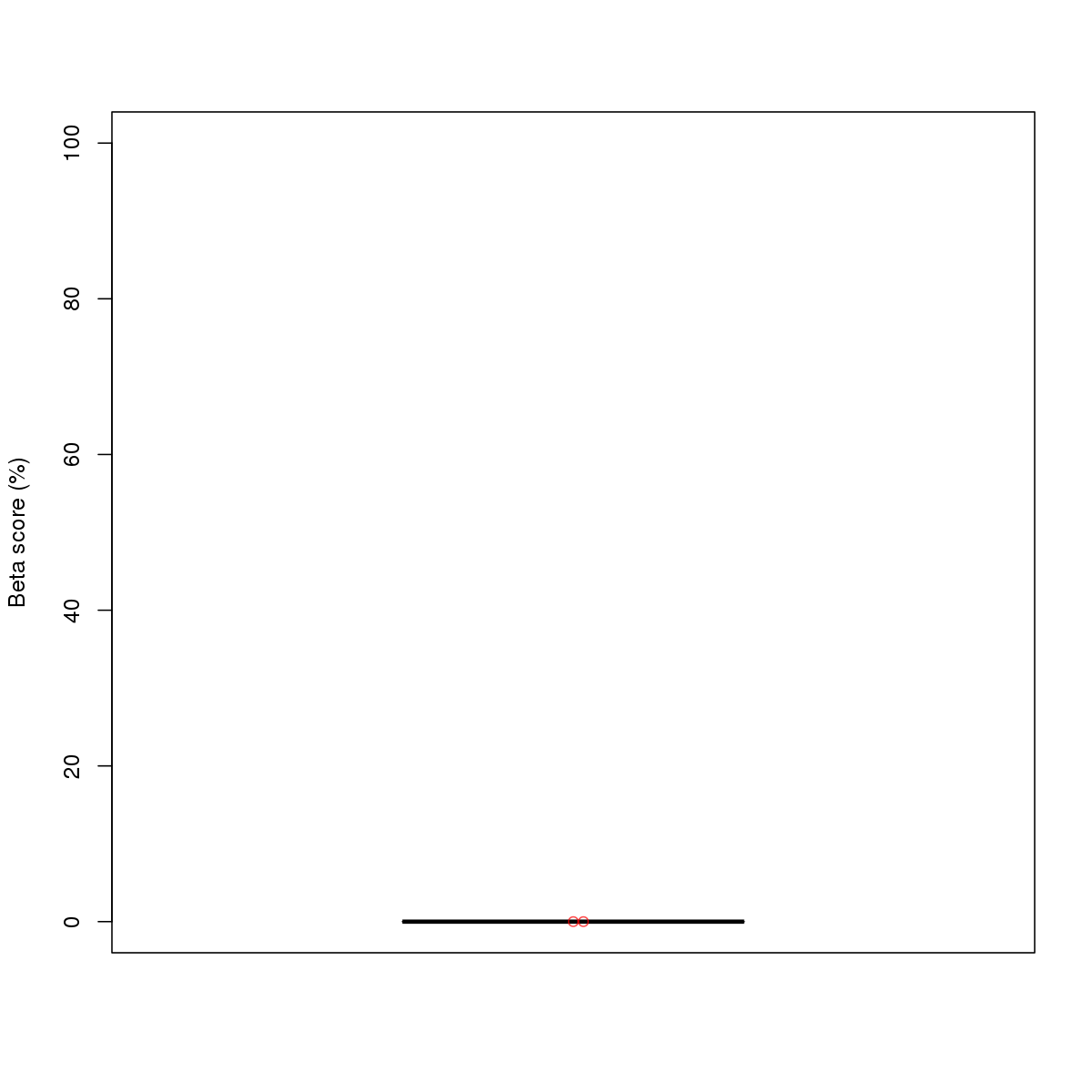 |  | Regions with USF1 motif: CG methScore in motif: Beta score matrix: |
| 10 | HCT116 | SRF (Details) | Responders to external signals (SRF/RLM1) | 5 | No CGs | 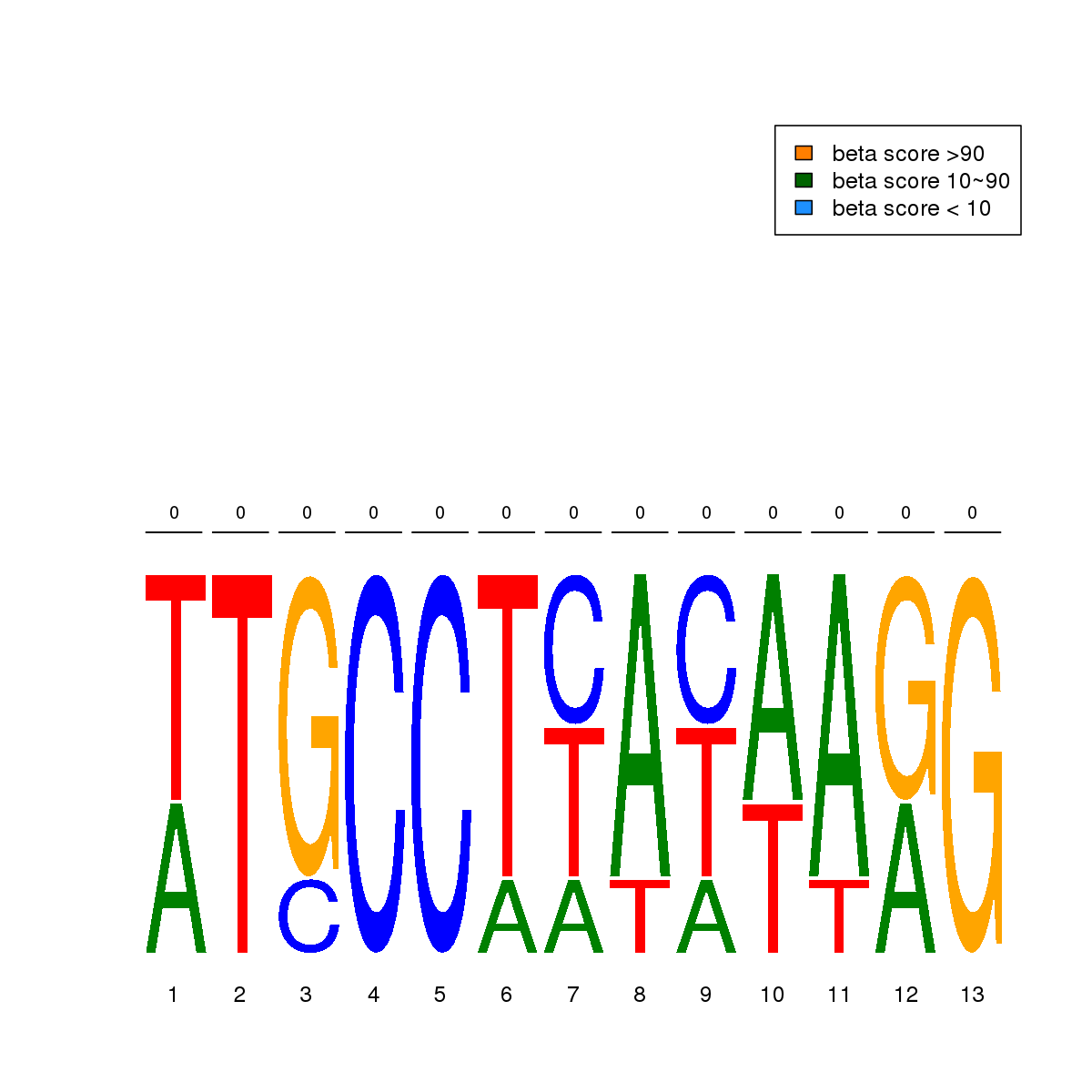 | Regions with SRF motif: CG methScore in motif: Beta score matrix: |
| 11 | HCT116 | YY1 (Details) | More than 3 adjacent zinc finger factors | 2 | No CGs | 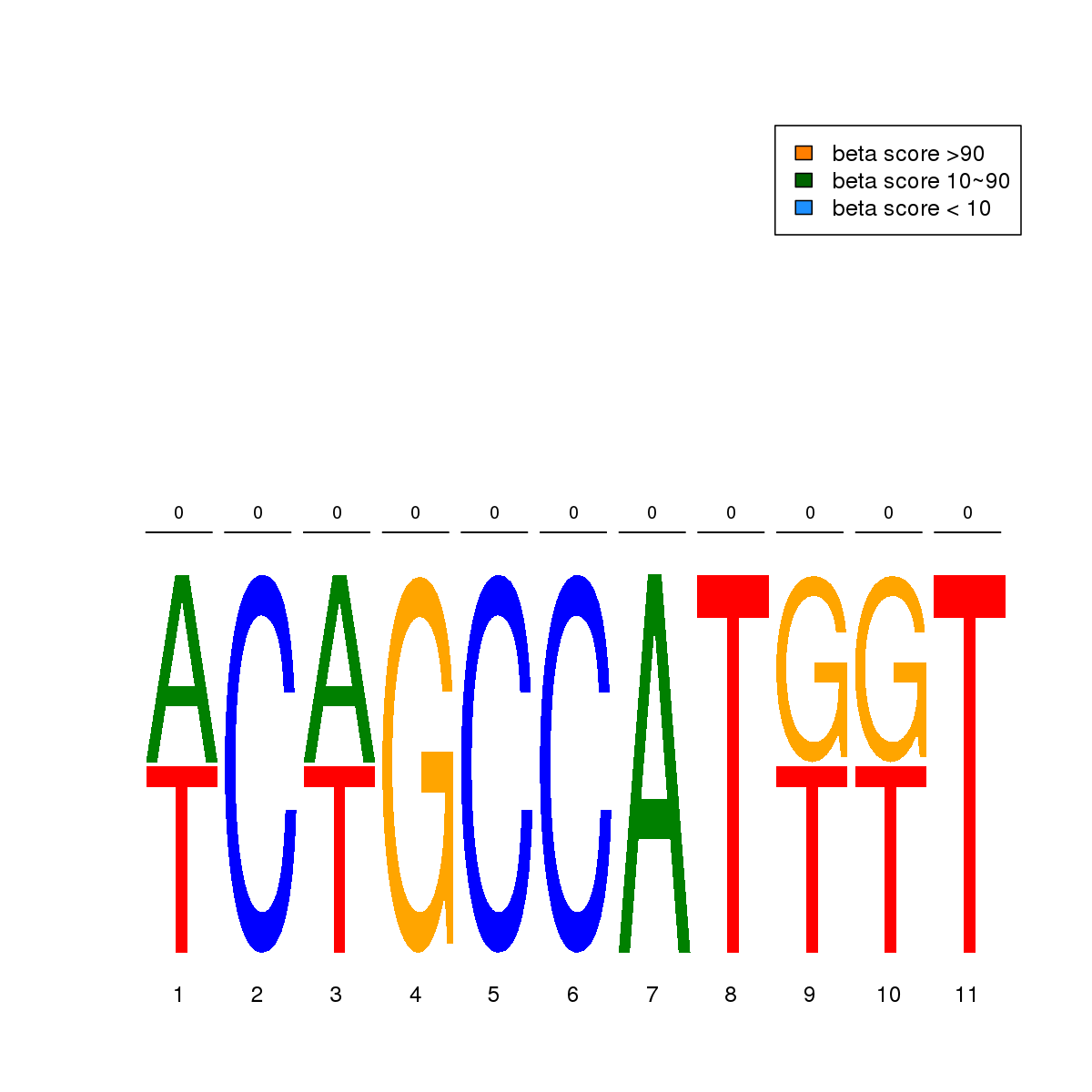 | Regions with YY1 motif: CG methScore in motif: Beta score matrix: |
| 12 | HCT116 | EGR1 (Details) | Three-zinc finger Kruppel-related factors | 1 | 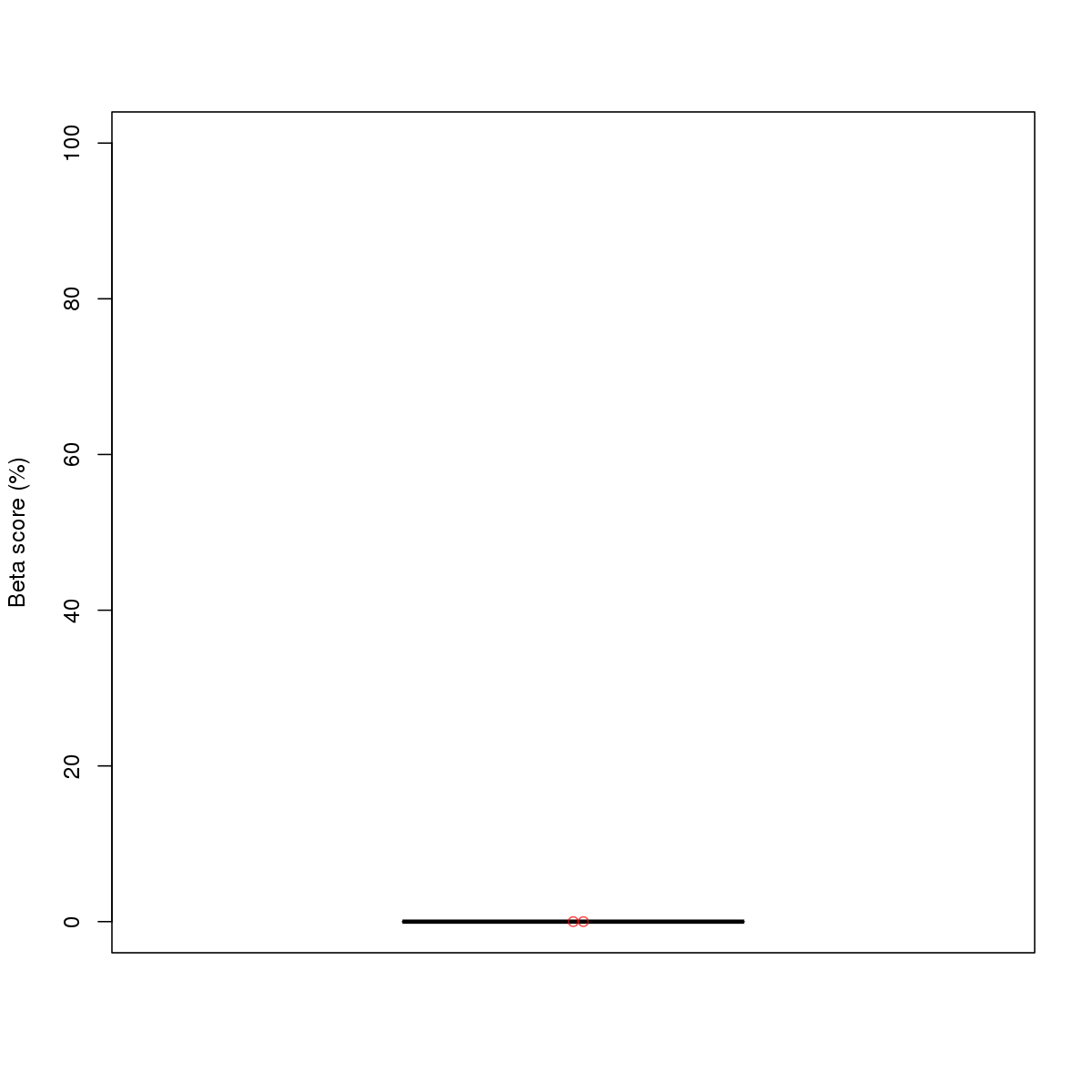 |  | Regions with EGR1 motif: CG methScore in motif: Beta score matrix: |
| 13 | HCT116 | MAX (Details) | bHLH-ZIP factors | 1 | No CGs |  | Regions with MAX motif: CG methScore in motif: Beta score matrix: |