
TFregulomeR is an R-library linking to a compendium of transcription factor (TF) cistrome and methylome data, derived from MethMotif and GTRD databases, while containing essential functions in R programming language to facilitate data browsing, retrieval and analysis. In particular, TFregulomeR enables the query of context in/dependent cistrome, TF interactome (global view of TF interactions at the DNA binding levels) as well as cis-regulatory modules. All these facilitate i) context specific co-factor analysis coupled with DNA methylation, ii) motif deconvolution and iii) annotation of context-specific TF binding sites in terms of genomic locations and functions.
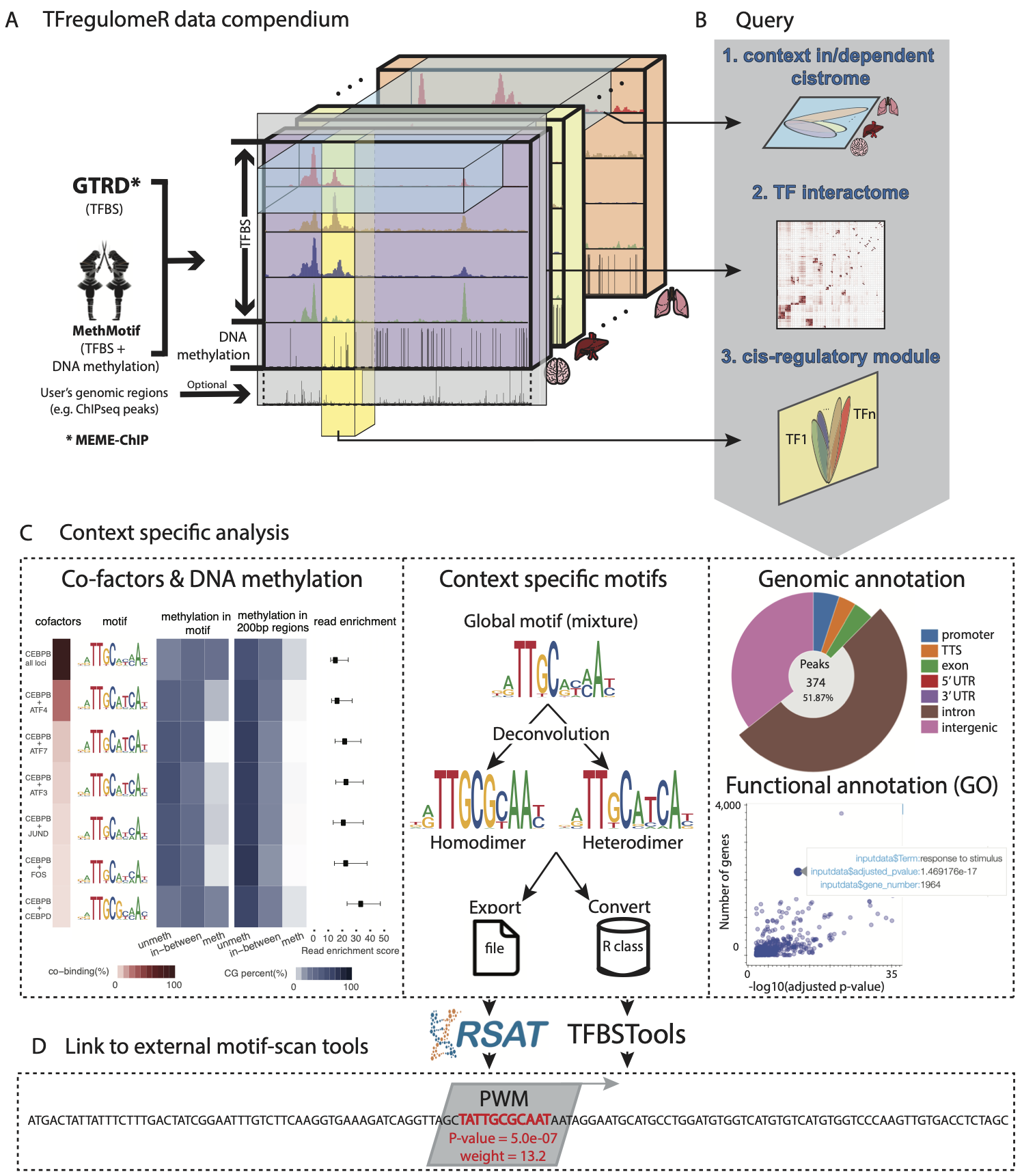
1) TFregulomeR linked data compendium
| item | Count |
|---|---|
| PWM | 1,468 |
| ChIP-seq experiments | 3,891 |
| Unique TF | 415 |
| PWM with DNA methylation records | 655 |
| Species | human (hg38) |
| Organ | stem_cell, blood_and_lymph, connective_tissue, colorectum, brain, bone, stomach, prostate, breast, pancreas, skin, kidney, lung, eye, esophagus, heart, muscle, uterus, spleen, cervix, testis, liver, adrenal_gland, neck_and_mouth, pleura, ovary, thymus, fallopian, vagina |
| Sample type | primary_cells, cell_line, tissue |
| Cell or tissue | 414 |
| Disease state | normal, tumor, Simpson_Golabi_Behmel_syndrome, progeria, metaplasia, unknown, immortalized, premetastatic |
| Source | GTRD, MethMotif |
2) TFregulomeR functionalities
TFregulomeR source codes have been implemented in GitHub.
1) For stable release repository, please visit here.
2) For development release repository, please visit here.
3) For a local version of the TFregulomeR database, please download and unzip the file found here.
Please click here for TFregulomeR manual.
Please click here for TFregulomeR vignettes.
Currently, TFregulomeR manuscript has been accepted in Nucleic Acids Research.
If you are using TFregulomeR, please cite us as follows:
Quy Xiao Xuan Lin, Denis Thieffry, Sudhakar Jha, Touati Benoukraf. (2019) TFregulomeR reveals transcription factors’ context-specific features and functions. Nucleic Acids Res., 10.1093/nar/gkz1088. [Manuscript]